
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,044,729 – 17,044,846 |
| Length | 117 |
| Max. P | 0.999755 |

| Location | 17,044,729 – 17,044,846 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 75.84 |
| Mean single sequence MFE | -33.13 |
| Consensus MFE | -21.48 |
| Energy contribution | -21.60 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.65 |
| SVM decision value | 4.01 |
| SVM RNA-class probability | 0.999755 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
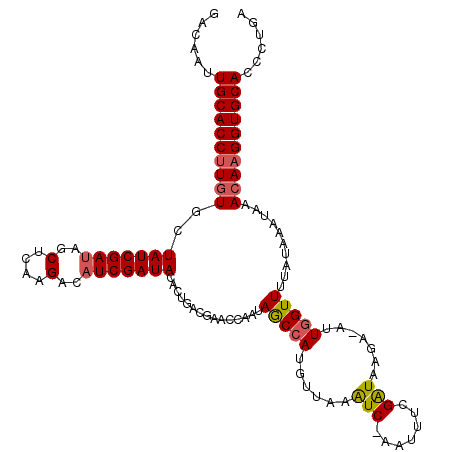
>3R_DroMel_CAF1 17044729 117 + 27905053 ACAGGGUGCACCUUGUGUAUUUAUAAAACCAAU-UCUAACCGAAAUU-GACUGAACAUGGUUAUUGCGAUGUCAGUAUAUCGAUGUCUUGAGCUAUCGAUAGCACAAGGUGCAAUUGUC ((((..((((((((((((...........((((-(........))))-)(((((.(((.((....)).)))))))).((((((((.(....).)))))))))))))))))))).)))). ( -41.10) >DroSim_CAF1 1254 118 + 1 UCAGGGUGCACCUUGUUUAUUUAUAAAACCAAU-UCUUAUCGAAAUUCGAUUUAACAUGGCUAUUGGCUCGUCAGUGUAUCGAUGUCUUGAGCUAUCGAUAGCACAAGGUGCAAUUGUC .(((..((((((((((.............((..-....((((.....))))......))((((((((((((.((.........))...)))))...))))))))))))))))).))).. ( -33.10) >DroYak_CAF1 1253 110 + 1 UCAGGGUGCACCUUGUUUAUUUAUAAAACUAAAUUUAUAUCGAUAAUAGAUUCGACAUGGCUAUCUAUCCACGCAUCUAUCGAU---------UGUCGAUAGCAUACGGUGCAAUUGUC .(((..((((((..((((.......))))........((((((((((((((.((...(((........))))).))))))....---------))))))))......)))))).))).. ( -25.20) >consensus UCAGGGUGCACCUUGUUUAUUUAUAAAACCAAU_UCUUAUCGAAAUU_GAUUCAACAUGGCUAUUGACACGUCAGUAUAUCGAUGUCUUGAGCUAUCGAUAGCACAAGGUGCAAUUGUC .(((..((((((((((.............................................(((((......)))))((((((((........))))))))..)))))))))).))).. (-21.48 = -21.60 + 0.12)



| Location | 17,044,729 – 17,044,846 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 75.84 |
| Mean single sequence MFE | -31.07 |
| Consensus MFE | -17.96 |
| Energy contribution | -18.72 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.58 |
| SVM decision value | 3.11 |
| SVM RNA-class probability | 0.998460 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17044729 117 - 27905053 GACAAUUGCACCUUGUGCUAUCGAUAGCUCAAGACAUCGAUAUACUGACAUCGCAAUAACCAUGUUCAGUC-AAUUUCGGUUAGA-AUUGGUUUUAUAAAUACACAAGGUGCACCCUGU .(((..(((((((((((.(((((((..(....)..))))))).((((((((.(......).))).)))))(-(((((......))-))))............)))))))))))...))) ( -32.50) >DroSim_CAF1 1254 118 - 1 GACAAUUGCACCUUGUGCUAUCGAUAGCUCAAGACAUCGAUACACUGACGAGCCAAUAGCCAUGUUAAAUCGAAUUUCGAUAAGA-AUUGGUUUUAUAAAUAAACAAGGUGCACCCUGA ......((((((((((..(((((((..(....)..))))))).......(((((((((((...)))).((((.....))))....-.))))))).........))))))))))...... ( -31.30) >DroYak_CAF1 1253 110 - 1 GACAAUUGCACCGUAUGCUAUCGACA---------AUCGAUAGAUGCGUGGAUAGAUAGCCAUGUCGAAUCUAUUAUCGAUAUAAAUUUAGUUUUAUAAAUAAACAAGGUGCACCCUGA ......((((((((((.((((((...---------..))))))))))((((((.((((....))))..))))))......((((((......)))))).........))))))...... ( -29.40) >consensus GACAAUUGCACCUUGUGCUAUCGAUAGCUCAAGACAUCGAUACACUGACGAACCAAUAGCCAUGUUAAAUC_AAUUUCGAUAAGA_AUUGGUUUUAUAAAUAAACAAGGUGCACCCUGA ......((((((((((..(((((((..(....)..)))))))...............(((((......(((.......))).......)))))..........))))))))))...... (-17.96 = -18.72 + 0.76)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:25 2006