
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,038,143 – 17,038,250 |
| Length | 107 |
| Max. P | 0.827330 |

| Location | 17,038,143 – 17,038,250 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.29 |
| Mean single sequence MFE | -47.98 |
| Consensus MFE | -30.80 |
| Energy contribution | -32.33 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827330 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
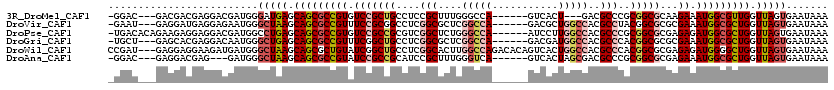
>3R_DroMel_CAF1 17038143 107 + 27905053 -GGAC---GACGACGAGGACGAUGGGAUGAGCAGCGCCGUGUCCGCUGCCUCCGCUUUGGGCCA------GUCACU---GACGCCCGCGGCGCAAGAAAUGGCGUUGGUUAGUGAAUAAA -....---.(((((..((.(((.((((.(.((((((.......)))))))))).).)))..)).------)))((.---.(((((.....(....)....)))))..))..))....... ( -36.10) >DroVir_CAF1 80271 110 + 1 -GAAU---GAGGAUGAGGAGAAUGGGCUAAGCAGCGCCGUUUCCGCGGCCUCGGCGCUCGGCCA------GACGCUGGCCACGCCUACGGCGCGCGAAAUGGCGCUGGUUAGUGAAUAAA -....---.................(((((.((((((((((((((((.((..((((...(((((------(...)))))).))))...)))))).)))))))))))).)))))....... ( -55.30) >DroPse_CAF1 57999 113 + 1 -UGACACAGAAGAGGAGGACGAUGGCCUGAGCAGCGCCGUGUCCGCCGCGUCGGCUCUGGGCCA------AUCCUUGGCCACGCCCGCGGCGCGAGAGAUGGCGCUGGUUAGUGAAUAAA -...(((........(((.(....))))((.(((((((((.(((((((((..(((....(((((------.....)))))..))))))))))...)).))))))))).)).)))...... ( -52.70) >DroGri_CAF1 65229 110 + 1 -UGCU---GAGCACGAGGACAAUGGGCUGAGCAGCGCCGUUUCGGCUGCCUCGGCGCUCGGCCA------GACGAUGGCCACGCCCACGGCGCGCGAAAUGGCGCUGGUUAGUGAAUAAA -....---.................(((((.(((((((((((((((.(((..((((...(((((------.....))))).))))...))))).))))))))))))).)))))....... ( -54.50) >DroWil_CAF1 74373 117 + 1 CCGAU---GAGGAGGAAGAUGAUGGGCUAAGCAGCGCUGUAUCGGCUGCCUCGGCACUUGGCCAGACACAGUCACUGGCCACGCCCACGGCGCGAGAGAUGGGGCUGGUUAGUGAAUAAA .....---((((.((..((((..(.(((....))).)..))))..)).))))(((.....)))........((((((((((.((((.((.(....)...))))))))))))))))..... ( -45.90) >DroAna_CAF1 59200 107 + 1 -GGAC---GAGGACGAG---GAUGGGCUAAGCAGCGCCGUAUCCGCCGCAUCCGCUUUGGGUCA------GUCACUAGCGACGCCCGCGGCGCGAGAAAUGGCGCUGGUUAGUGAAUAAA -....---.........---.....(((((.(((((((((.((((((((.........((((..------(((......))))))))))))).))...))))))))).)))))....... ( -43.40) >consensus _GGAC___GAGGACGAGGACGAUGGGCUAAGCAGCGCCGUAUCCGCUGCCUCGGCUCUGGGCCA______GUCACUGGCCACGCCCACGGCGCGAGAAAUGGCGCUGGUUAGUGAAUAAA .........................(((((.(((((((((.(((((((....(((....(((((...........)))))..)))..)))))...)).))))))))).)))))....... (-30.80 = -32.33 + 1.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:21 2006