| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,037,453 – 17,037,560 |
| Length | 107 |
| Max. P | 0.664120 |

| Location | 17,037,453 – 17,037,560 |
|---|---|
| Length | 107 |
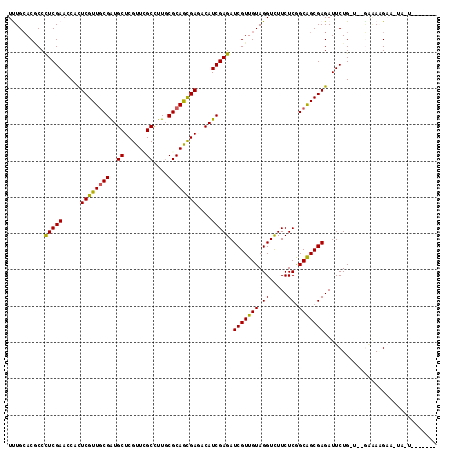
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 81.45 |
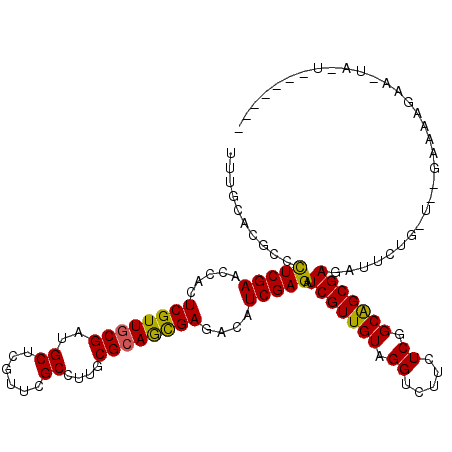
| Mean single sequence MFE | -36.48 |
| Consensus MFE | -26.45 |
| Energy contribution | -25.95 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.664120 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17037453 107 - 27905053 UCUGCACGCCCUCGAACCACUCGUUCCGGUGCUCGUUGGCUUUGCGCAGCGAAACAUCGAGAUCGUUGUAGGUUUUCUCGGCGGCGAGAUUCUG-AACGAAAAGAA-UA--------- ..(((.((((...(((((((.((.(((((((.((((((((...)).))))))..))))).)).))..)).)))))....)))))))..(((((.-.......))))-).--------- ( -35.10) >DroVir_CAF1 79515 113 - 1 UUUGGACGCCCUCGAACCACUCGUUGCGAUGCUCGUUCGCCUUGCGCAGCGAGACAUCGAGAUCGUUGUAGGUCUUCUCGGCAGCGAGAUUCUG-U--UAAAGGAUUAA-GACACAG- .........(((..(((..(((((((((((((((((((((...))).)))))).))))((((((......))))))....)))))))).....)-)--)..))).....-.......- ( -38.80) >DroGri_CAF1 64441 106 - 1 UUUGCACACCCUCGAACCACUCGUUGCGAUGCUCGUUCGCCUUGCGCAGCGAGACAUCGAGAUCGUUGUAGGUCUUCUCGGCAGCGAGAUUCUG-U--GAGAAGAA-UA-U------- ...................(((((((((((((((((((((...))).)))))).))))((((((......))))))....))))))))(((((.-.--....))))-).-.------- ( -38.60) >DroWil_CAF1 73662 117 - 1 UUUGCACACCUUCGAACCAUUCGUUGCGAUGCUCGUUUGCCUUUCGCAAUGAUACAUCGAGAUCGUUGUAGGUUUUCUCAGCGGCGAGAUUCUGCAAUGCAAUGCA-UAAUAAAAAAC ..((((....(((((.....(((((((((.((......))...)))))))))....)))))..(((((((((...((((......)))).)))))))))...))))-........... ( -35.60) >DroMoj_CAF1 66288 112 - 1 UCUGCACGCCCUCGAACCACUCGUUGCGAUGCUCGUUCGCCUUGCGCAGCGAGACAUCGAGAUCGUUGUAGGUCUUCUCAGCAGCGAGAUUCUG-U--GGAA-GAUAAA-UGCAAUG- ..((((.............(((((((((((((((((((((...))).)))))).))))((((((......))))))....))))))))..(((.-.--...)-))....-))))...- ( -40.00) >DroAna_CAF1 58462 104 - 1 UUUGCACCCCCUCGAACCACUCGUUGCGAUGCUCGUUAGCCUUGCGCAGCGACACAUCGAGAUCGUUGUAGGUUUUCUCGGCGGCGAGAUUCUG-UAAAAUAGGC------------- ((((((....(((((.....((((((((..(((....)))....))))))))....))))).(((((((.((.....)).))))))).....))-))))......------------- ( -30.80) >consensus UUUGCACGCCCUCGAACCACUCGUUGCGAUGCUCGUUCGCCUUGCGCAGCGAGACAUCGAGAUCGUUGUAGGUCUUCUCGGCAGCGAGAUUCUG_U__GAAAAGAA_UA_U_______ ..........(((((.....((((((((..((......))....))))))))....))))).(((((((.((.....)).)))))))............................... (-26.45 = -25.95 + -0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:20 2006