
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 17,004,765 – 17,004,879 |
| Length | 114 |
| Max. P | 0.795201 |

| Location | 17,004,765 – 17,004,879 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 80.02 |
| Mean single sequence MFE | -35.42 |
| Consensus MFE | -25.83 |
| Energy contribution | -25.33 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598903 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
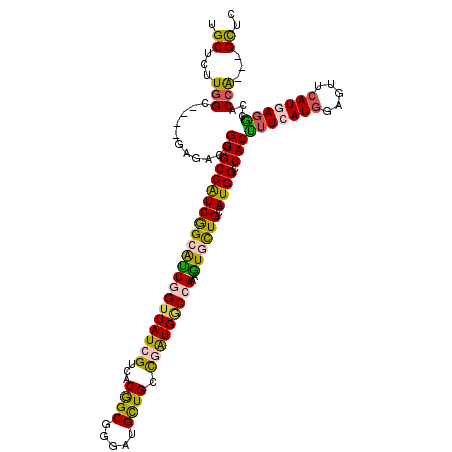
>3R_DroMel_CAF1 17004765 114 + 27905053 GAGC---CGGUGGCCUUAUGAACUCCAUGAAGGUGGCCAUGCAGAACUUUAGCCAUCGCCAGCAUCCCGCCGUGACGAUAACGAGCGCCGACGGCACGCAAUCAACGGCAAAGAGCA ..((---((.((((.(((((.....))))).((((((..............))))))))))((.....(((((.......))).))(((...)))..))......))))........ ( -34.24) >DroVir_CAF1 38734 111 + 1 GCGC---CGGCGGCCUCAUGAACUCCAUGAAAGUGGCCAUGCAGCAUUUUGGCCAUAAACAACAUCCCGCCGUAACGAUAACCAAUGCCGAUGGCACCGUCUC---CGCCAAGAGCA .((.---((((((..(((((.....)))))..(((((((..........)))))))..........))))))...))........((((..((((........---.)))).).))) ( -33.50) >DroGri_CAF1 29631 114 + 1 GUGCUGGUGGCGGUCUCAUGAAUUCCAUGAAAGUGGCCAUGCAACACUUUGGCCAUCGACAGCAUCCCGCCGUAACGAUAACCAAUGCCGAUGGCACCGUCUC---CGCCAAGAGCA .((((..((((((..(((((.....))))).....((((((((.....((((..((((((.((.....)).))..))))..)))))))..))))).......)---)))))..)))) ( -37.90) >DroWil_CAF1 37350 111 + 1 CAAC---UGGUGGCAUCAUGAACUCUAUGAAAGUGGCCAUGCAACACUUUGGACACCGGCAACAUCCCGCUGUAACGAUAACCAAUGCCGAUGGCACUGUCU---CGGCCAAAAGCA ....---((((.((.(((((.....)))))..)).))))(((......(((((((.(((.......))).)))........)))).(((((.(((...))))---)))).....))) ( -25.90) >DroMoj_CAF1 31226 105 + 1 GCAC---UGGCGGCCUCAUGAAUUCCAUGAAAGUGGCCAUGCAGCACUUUGGUCAUAAGCAACAUCCCGCCGUGACGAUAACCAGUGCUGAUGGCACC---------GCCAAGAGCA ((.(---((((((..(((((.....))))).....(((((.(((((((.(.(((((..((........)).))))).).....)))))))))))).))---------))).)).)). ( -42.60) >DroAna_CAF1 26064 111 + 1 GAGC---CGGUGGCCUUAUGAACUCCAUGAAGGUGGCCAUGCAGAACUUUGGCCACCGCCAGCACCCCGCCGUGACGAUAACUAACGCCGACGGCAC--AGUCUCC-GCCAAGAGCA (((.---((.((((.(((((.....))))).((((((((..........)))))))))))).).....((((((.((........)).).)))))..--.).))).-((.....)). ( -38.40) >consensus GAGC___CGGCGGCCUCAUGAACUCCAUGAAAGUGGCCAUGCAGCACUUUGGCCAUCGACAACAUCCCGCCGUAACGAUAACCAAUGCCGAUGGCACCGUCUC____GCCAAGAGCA .......((((((..(((((.....)))))..(((((((..........)))))))..........))))))..............((...((((............))))...)). (-25.83 = -25.33 + -0.50)



| Location | 17,004,765 – 17,004,879 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 80.02 |
| Mean single sequence MFE | -43.72 |
| Consensus MFE | -30.75 |
| Energy contribution | -29.82 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795201 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 17004765 114 - 27905053 UGCUCUUUGCCGUUGAUUGCGUGCCGUCGGCGCUCGUUAUCGUCACGGCGGGAUGCUGGCGAUGGCUAAAGUUCUGCAUGGCCACCUUCAUGGAGUUCAUAAGGCCACCG---GCUC (((.(((((((((((...((((.((((((..((........))..)))))).))))...))))))).))))....))).((((.((((.(((.....))))))).....)---))). ( -41.60) >DroVir_CAF1 38734 111 - 1 UGCUCUUGGCG---GAGACGGUGCCAUCGGCAUUGGUUAUCGUUACGGCGGGAUGUUGUUUAUGGCCAAAAUGCUGCAUGGCCACUUUCAUGGAGUUCAUGAGGCCGCCG---GCGC .(((...((((---(....(((((((((((((((((((((....(((((.....)))))..)))))))..)))))).)))).))))((((((.....)))))).))))))---)).. ( -43.20) >DroGri_CAF1 29631 114 - 1 UGCUCUUGGCG---GAGACGGUGCCAUCGGCAUUGGUUAUCGUUACGGCGGGAUGCUGUCGAUGGCCAAAGUGUUGCAUGGCCACUUUCAUGGAAUUCAUGAGACCGCCACCAGCAC ((((..(((((---(.....((((((((((((((((((((((..(((((.....))))))))))))))..)))))).)))).)))(((((((.....)))))))))))))..)))). ( -52.80) >DroWil_CAF1 37350 111 - 1 UGCUUUUGGCCG---AGACAGUGCCAUCGGCAUUGGUUAUCGUUACAGCGGGAUGUUGCCGGUGUCCAAAGUGUUGCAUGGCCACUUUCAUAGAGUUCAUGAUGCCACCA---GUUG .(((..((((..---.((.(((((((((((((((((.(((((...((((.....)))).))))).)))..)))))).)))).)))).))...(....).....))))..)---)).. ( -31.70) >DroMoj_CAF1 31226 105 - 1 UGCUCUUGGC---------GGUGCCAUCAGCACUGGUUAUCGUCACGGCGGGAUGUUGCUUAUGACCAAAGUGCUGCAUGGCCACUUUCAUGGAAUUCAUGAGGCCGCCA---GUGC .((.((.(((---------(((((((((((((((((((((.((.(((......))).))..))))))..))))))).)))))....((((((.....)))))))))))))---).)) ( -45.20) >DroAna_CAF1 26064 111 - 1 UGCUCUUGGC-GGAGACU--GUGCCGUCGGCGUUAGUUAUCGUCACGGCGGGGUGCUGGCGGUGGCCAAAGUUCUGCAUGGCCACCUUCAUGGAGUUCAUAAGGCCACCG---GCUC .(((...(.(-((..(((--.((((((.((((........)))))))))).))).))).)(((((((...(..((.(((((......))))).))..)....))))))))---)).. ( -47.80) >consensus UGCUCUUGGC____GAGACGGUGCCAUCGGCAUUGGUUAUCGUCACGGCGGGAUGCUGCCGAUGGCCAAAGUGCUGCAUGGCCACUUUCAUGGAGUUCAUGAGGCCACCA___GCUC .((...(((...........((((((((((((((((((((((...((((.....)))).)))))))))..)))))).)))).)))(((((((.....)))))))...)))...)).. (-30.75 = -29.82 + -0.94)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:08 2006