
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,949,772 – 16,949,867 |
| Length | 95 |
| Max. P | 0.803791 |

| Location | 16,949,772 – 16,949,867 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 83.55 |
| Mean single sequence MFE | -29.25 |
| Consensus MFE | -19.00 |
| Energy contribution | -20.37 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.18 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.803791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
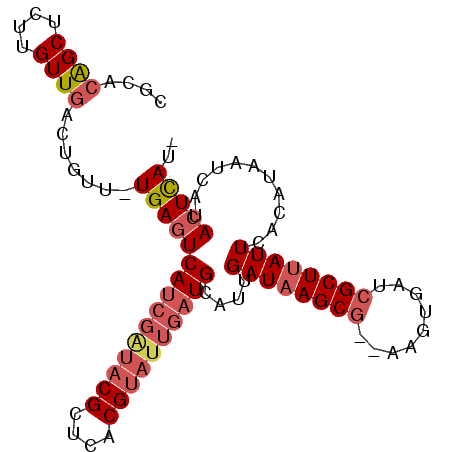
>3R_DroMel_CAF1 16949772 95 + 27905053 CGCACAGCUCUUGUUGAUUGUU-UGAGUCAUCGAUACGCUCACGUAUUGAUGCAUUGAUAAGCG--AACUGAUCGCUUAUUCACAUACUCAUACUCAU- ....((((....))))...((.-(((((((((((((((....))))))))))....((((((((--(.....))))))))).....))))).))....- ( -29.80) >DroSec_CAF1 2964 97 + 1 CGCACAGCUCUUGUUGGCUGUU-UGAGUCAUCGAUACGCUCACGUAUUGAUGCAUUGAUAAGCGAGAAGUGAUCGCUUAUUCACAUACUCAUACUCAU- .((.((((....)))))).((.-(((((((((((((((....))))))))))....(((((((((.......))))))))).....))))).))....- ( -33.10) >DroSim_CAF1 3004 97 + 1 CGCACAGCUCUUGUUGGCUGUU-UGAGUCAUCGAUACGCUCACGUAUUGAUGCAUUGAUAAGCGAGAAGUGAUCGCUUAUUCACAUACUCAUACUCAU- .((.((((....)))))).((.-(((((((((((((((....))))))))))....(((((((((.......))))))))).....))))).))....- ( -33.10) >DroEre_CAF1 2960 94 + 1 CGCACAGCU-UUGUUGACUGUU-UGAGUCAUCGAUACGCUCACGUAUUGAUGCAUUGAUAAGCG--AAGUGAUCGCUUAUUCACUAAAGCCGACUUAU- ....(.(((-(((.(((.....-...(.((((((((((....)))))))))))....(((((((--(.....))))))))))).)))))).)......- ( -28.50) >DroYak_CAF1 2954 95 + 1 CGCACAGCUCUUGUUGACUGUU-UGAGUCAUCGGUACGCUCACGUAUUGAUGCAUUGAUAAGCG--AAGUGAUCGCUUAUUCACUGAAGCCGACUCAU- ...((((.((.....)))))).-(((((((((((((((....))))))))).((.(((((((((--(.....))))))).))).)).....)))))).- ( -31.80) >DroAna_CAF1 2356 83 + 1 CGCACGGCUCUUGUUUACCAUUCUGAGUCAUCAGUA--------------UGCAUUGAUAAGCG--ACCUGAUGGCUGAUUCACAUAAUCCCAGUCAGC .((..(((.((((((...(((.((((....)))).)--------------))....)))))).)--.))...((((((.............)))))))) ( -19.22) >consensus CGCACAGCUCUUGUUGACUGUU_UGAGUCAUCGAUACGCUCACGUAUUGAUGCAUUGAUAAGCG__AAGUGAUCGCUUAUUCACAUAAUCAUACUCAU_ ....((((....)))).......(((((((((((((((....))))))))))....((((((((.........))))))))...........))))).. (-19.00 = -20.37 + 1.36)

| Location | 16,949,772 – 16,949,867 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 83.55 |
| Mean single sequence MFE | -27.90 |
| Consensus MFE | -18.84 |
| Energy contribution | -20.12 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.691526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
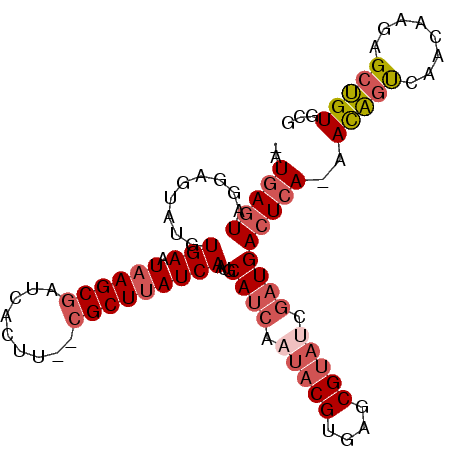
>3R_DroMel_CAF1 16949772 95 - 27905053 -AUGAGUAUGAGUAUGUGAAUAAGCGAUCAGUU--CGCUUAUCAAUGCAUCAAUACGUGAGCGUAUCGAUGACUCA-AACAAUCAACAAGAGCUGUGCG -....((.(((((...(((.(((((((.....)--)))))))))...((((.(((((....))))).)))))))))-.))................... ( -26.90) >DroSec_CAF1 2964 97 - 1 -AUGAGUAUGAGUAUGUGAAUAAGCGAUCACUUCUCGCUUAUCAAUGCAUCAAUACGUGAGCGUAUCGAUGACUCA-AACAGCCAACAAGAGCUGUGCG -.(((((....((((.(((.(((((((.......))))))))))))))(((.(((((....))))).))).)))))-.(((((........)))))... ( -29.90) >DroSim_CAF1 3004 97 - 1 -AUGAGUAUGAGUAUGUGAAUAAGCGAUCACUUCUCGCUUAUCAAUGCAUCAAUACGUGAGCGUAUCGAUGACUCA-AACAGCCAACAAGAGCUGUGCG -.(((((....((((.(((.(((((((.......))))))))))))))(((.(((((....))))).))).)))))-.(((((........)))))... ( -29.90) >DroEre_CAF1 2960 94 - 1 -AUAAGUCGGCUUUAGUGAAUAAGCGAUCACUU--CGCUUAUCAAUGCAUCAAUACGUGAGCGUAUCGAUGACUCA-AACAGUCAACAA-AGCUGUGCG -....(((((((((..(((((((((((.....)--))))))).....((((.(((((....))))).)))).....-.....)))..))-))))).)). ( -30.20) >DroYak_CAF1 2954 95 - 1 -AUGAGUCGGCUUCAGUGAAUAAGCGAUCACUU--CGCUUAUCAAUGCAUCAAUACGUGAGCGUACCGAUGACUCA-AACAGUCAACAAGAGCUGUGCG -.((((((.((.....(((.(((((((.....)--)))))))))..))(((..((((....))))..)))))))))-.(((((........)))))... ( -29.60) >DroAna_CAF1 2356 83 - 1 GCUGACUGGGAUUAUGUGAAUCAGCCAUCAGGU--CGCUUAUCAAUGCA--------------UACUGAUGACUCAGAAUGGUAAACAAGAGCCGUGCG ((...(((((.....((......))((((((..--.((........)).--------------..)))))).))))).(((((........))))))). ( -20.90) >consensus _AUGAGUAGGAGUAUGUGAAUAAGCGAUCACUU__CGCUUAUCAAUGCAUCAAUACGUGAGCGUAUCGAUGACUCA_AACAGUCAACAAGAGCUGUGCG ..(((((.........(((.((((((.........)))))))))...((((.(((((....))))).)))))))))..(((((........)))))... (-18.84 = -20.12 + 1.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:00 2006