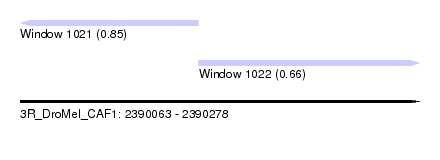
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,390,063 – 2,390,278 |
| Length | 215 |
| Max. P | 0.846359 |

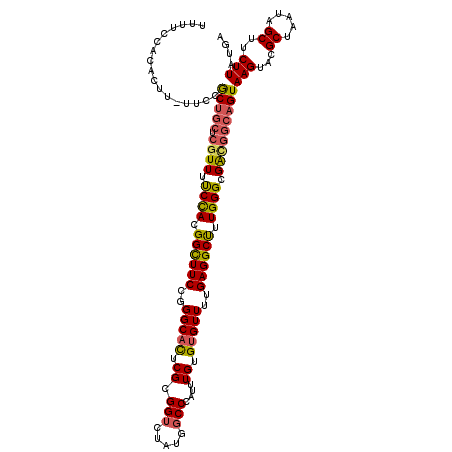
| Location | 2,390,063 – 2,390,159 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 80.05 |
| Mean single sequence MFE | -23.87 |
| Consensus MFE | -14.04 |
| Energy contribution | -14.77 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846359 |
| Prediction | RNA |
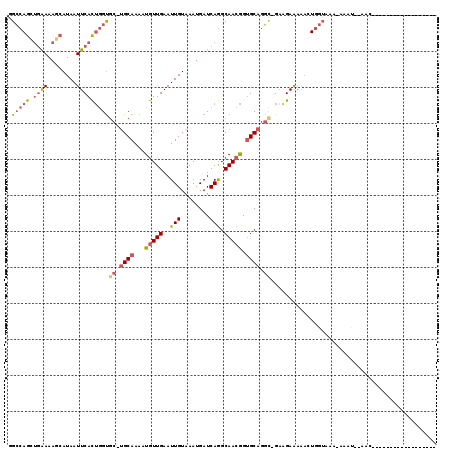
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2390063 96 - 27905053 GGCCAGUUGAAAAGCAUAAUUCACUGGUGC-UGCAAAAUGUUGAAUUGUAAAUGAUCAGGCAACGGUGCAGGC-GAGGAAAAACUGGUAAA-AAAU--AAC------------------ .((((((((((........))).((.((.(-((((...(((((..(((........))).))))).)))))))-.))....)))))))...-....--...------------------ ( -24.90) >DroGri_CAF1 164261 116 - 1 GGAAAACAUGAAAACAUAAUUCAUGAGCUCGAGCAAAAUGUUGAAUUGUAAAUGAUCAAGCAAGAG-GCGGUCUGAAGAGGG-AUGGGGCA-AGAACCAGCUACAGAGACAGCUACAGG ((....((((((.......)))))).((((..((....((((..(((......)))..))))....-)).((((......))-)).)))).-....))((((........))))..... ( -21.80) >DroSec_CAF1 103338 97 - 1 GGCCAGCUGAAAAGCAUAAUUCACUGGUGC-UGCAAAAUGUUGAAUUGUAAAUGAUCAGGCAACGGUGCAGGC-GAGGAAAAACUGGUAAAAAAAC--AAC------------------ .(((((((....)))....(((.((.((.(-((((...(((((..(((........))).))))).)))))))-.)))))....))))........--...------------------ ( -26.20) >DroSim_CAF1 95354 97 - 1 GGCCAGCUGAAAAGCAUAAUUCACUGGUGC-UGCAAAAUGUUGAAUUGUAAAUGAUCAGGCAACGGUGCAGGC-GAGGAAAAACUGGUAAAAAAAU--AAC------------------ .(((((((....)))....(((.((.((.(-((((...(((((..(((........))).))))).)))))))-.)))))....))))........--...------------------ ( -26.20) >DroEre_CAF1 97023 96 - 1 GGCCAGCUGAAAAGCAUAAUUCACUGGUGC-UGCAAAAUGUUGAAUUGUAAAUGAUCAGGCAACGGUGCAGGU-GUAGAAAAAAUGGUACA-AAAU--AAA------------------ .(((((((....)))........(((...(-((((...(((((..(((........))).))))).)))))..-.)))......))))...-....--...------------------ ( -21.70) >DroYak_CAF1 91144 96 - 1 GGCCAGCUGAAAAGCAUAAUUCACUGGUGC-UGCAAAAUGUUGAAUUGUAAAUGAUCAGGCAACGGUGCAGGC-GAAGAAAAAGUGGUACA-AAAU--AAC------------------ .(((((((....)))........((..(((-((((...(((((..(((........))).))))).)))).))-).))......))))...-....--...------------------ ( -22.40) >consensus GGCCAGCUGAAAAGCAUAAUUCACUGGUGC_UGCAAAAUGUUGAAUUGUAAAUGAUCAGGCAACGGUGCAGGC_GAAGAAAAACUGGUAAA_AAAU__AAC__________________ .(((((.((((........)))))))))((.((((...(((((..(((........))).))))).)))).)).............................................. (-14.04 = -14.77 + 0.73)

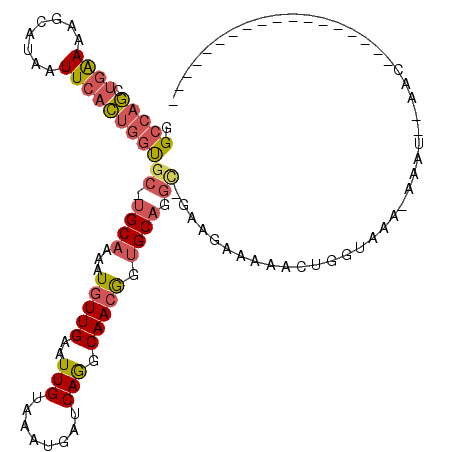
| Location | 2,390,159 – 2,390,278 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.34 |
| Mean single sequence MFE | -37.87 |
| Consensus MFE | -33.38 |
| Energy contribution | -33.38 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2390159 119 + 27905053 UUUUCCCCACUU-UUCCGCUGCUCGUUUUCCACGGCUUCCGGGCACUCGCGGUCUAUGGCCCAUUUGUGUGUUUUGAGGCUUUGGGCGAUGGCAGUAAGUACGCUAAUAGCUUCUUAUGA ........((((-....(((((.((((.((((.((((((..(((((.((.(((.....)))....)).)))))..)))))).)))).)))))))))))))..((.....))......... ( -36.50) >DroSec_CAF1 103435 119 + 1 UUUUCCCCACUU-UUCCGCUGCUCGUUUUCCACGGCUUCCGGGCACUCGCGGUCUAUGGCCCAUUUGUGUGUUUUGAGGCUUUGGGCGGCGGCAGUAAGUACGCUAAUAGCUUCUUAUGA ........((((-....(((((.(((..((((.((((((..(((((.((.(((.....)))....)).)))))..)))))).))))..))))))))))))..((.....))......... ( -38.30) >DroSim_CAF1 95451 119 + 1 UUUUCCCCACUU-UUCCGCUGCUCGUUUUCCACGGCUUCCGGGCACUCGCGGUCUAUGGCCCAUUUGUGUGUUUUGAGGCUUUGGGCGACGGCAGUAAGUACGCUAAUAGCUUCUUAUGA ........((((-....(((((.((((.((((.((((((..(((((.((.(((.....)))....)).)))))..)))))).)))).)))))))))))))..((.....))......... ( -38.30) >DroEre_CAF1 97119 119 + 1 UUUUCCACACUU-UCCCACUGCUCGUUUUCCACGGCUUCCGGGCAUUCGCGGUCUAUGGCCCAUUUGUGUGUUUUGAGGCCUUGGGCGACGGCAGUAAGUACGCUAAUAGCUUCUUAUGA ........((((-....(((((.((((.((((.((((((..(((((.((.(((.....)))....)).)))))..)))))).)))).)))))))))))))..((.....))......... ( -37.60) >DroYak_CAF1 91240 120 + 1 UUUUCCACACUUUUCCCGCUGCUCGUUUUCCACGGCUUCCGGGCACUCGCGGUCUAUGGCCCAUUUGUGUGUUUUGAGGCCUUGGGCGACGGCAGUAAGUACGCUAAUAGCUUCUUAUGA .................(((((.((((.((((.((((((..(((((.((.(((.....)))....)).)))))..)))))).)))).)))))))))(((...((.....))..))).... ( -39.80) >DroAna_CAF1 95337 119 + 1 GUUUCUAUGCCA-UUUUGCAAGGCAUUUCCUAUGGUUUCCGGGCACUCGCGGGCCAUGGCCCAUUUGUGGGUUUUGAGGCUUUGGGCGACAGCAGUAAGUACGCUAAUAGCUUCUUAUGA ......(((((.-........)))))...((((((((..(((....)))..))))))))((((....))))....((((((...((((((........)).))))...))))))...... ( -36.70) >consensus UUUUCCACACUU_UUCCGCUGCUCGUUUUCCACGGCUUCCGGGCACUCGCGGUCUAUGGCCCAUUUGUGUGUUUUGAGGCUUUGGGCGACGGCAGUAAGUACGCUAAUAGCUUCUUAUGA .................(((((.((((.((((.((((((..(((((.((.(((.....)))....)).)))))..)))))).)))).)))))))))(((...((.....))..))).... (-33.38 = -33.38 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:14 2006