
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,857,558 – 16,857,718 |
| Length | 160 |
| Max. P | 0.998416 |

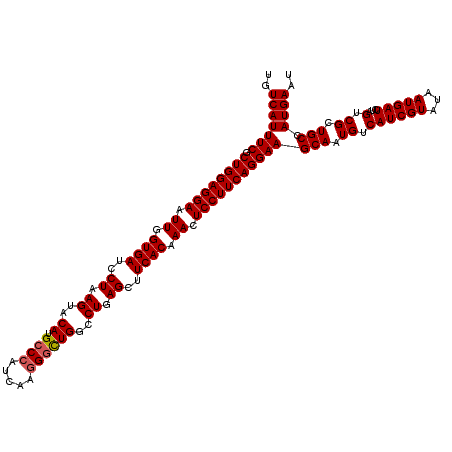
| Location | 16,857,558 – 16,857,678 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -34.95 |
| Consensus MFE | -32.27 |
| Energy contribution | -32.35 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.02 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924363 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
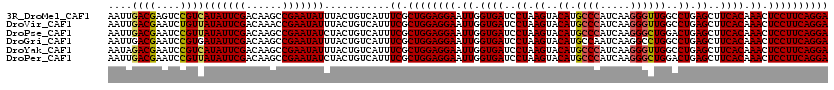
>3R_DroMel_CAF1 16857558 120 - 27905053 UGUCAUUUCGCUGGAGGAAUUGGUGAUCCUAAGUACAUGCCCAUCAAGGGUUGGCCUGAGCUUCACAAACUCCUUCAGGAAGCAAUGUCAUCGUAUAAUGAUCUAGUCGCUGCCAUGAAC ..(((((((.((((((((.((.((((..((.((..((.((((.....))))))..)).))..)))).)).)))))))))))(((.((.((((((...)))))...).)).))).)))).. ( -34.00) >DroVir_CAF1 11098 120 - 1 UGUCAUUUCGCUGGAGGAAUUGGUGAUCCUAAGUACAUGCCCAUCAAGGGUUGGCCUGAGCUUCACAAACUCCUUCAGGAAGCAAUGUCAUCGUAUAAUGAUCUGGUCGCUGCCAUGAAU ..(((((((.((((((((.((.((((..((.((..((.((((.....))))))..)).))..)))).)).)))))))))))(((.(((((((((...))))..))).)).))).)))).. ( -34.40) >DroPse_CAF1 18902 120 - 1 UGUCAUUUCGCUGGAGGAAUUGGUGAUCCUAAGUACAUGCCCAUCAAGGGCUGGACUGAGCUUCACAAACUCCUUCAGGAAGCAAUGUCAUCGUAUAAUGAUCUAGUCGCUGCCAUGAAU ..(((((((.((((((((.((.((((..((.(((.((.((((.....)))))).))).))..)))).)).)))))))))))(((.((.((((((...)))))...).)).))).)))).. ( -36.70) >DroGri_CAF1 12733 120 - 1 UGUCAUUUCGCUGGAGGAAUUGGUGAUCCUAAGUACAUGCCAAUCAAGGCCUGGCCUGAGCUUCACAAACUCCUUCAGGAAGCAAUGUCAUCGUAUAAUGAUCUAGUCGCUGCCAUGAAU ..(((((((.((((((((.((.((((..((.((..((.(((......))).))..)).))..)))).)).)))))))))))(((.((.((((((...)))))...).)).))).)))).. ( -33.90) >DroYak_CAF1 12566 120 - 1 UGUCAUUUCGCUGGAGGAAUUGGUGAUCCUAAGUACAUGCCCAUCAAGGGUUGGCCUGAGCUUCACAAACUCCUUCAGGAAGCAAUGUCAUCGUAUAAUGAUCUAGUCGCUGCCAUGAAC ..(((((((.((((((((.((.((((..((.((..((.((((.....))))))..)).))..)))).)).)))))))))))(((.((.((((((...)))))...).)).))).)))).. ( -34.00) >DroPer_CAF1 19505 120 - 1 UGUCAUUUCGCUGGAGGAAUUGGUGAUCCUAAGUACAUGCCCAUCAAGGGCUGGACUGAGCUUCACAAACUCCUUCAGGAAGCAAUGUCAUCGUAUAAUGAUCUAGUCGCUGCCAUGAAU ..(((((((.((((((((.((.((((..((.(((.((.((((.....)))))).))).))..)))).)).)))))))))))(((.((.((((((...)))))...).)).))).)))).. ( -36.70) >consensus UGUCAUUUCGCUGGAGGAAUUGGUGAUCCUAAGUACAUGCCCAUCAAGGGCUGGCCUGAGCUUCACAAACUCCUUCAGGAAGCAAUGUCAUCGUAUAAUGAUCUAGUCGCUGCCAUGAAU ..(((((((.((((((((.((.((((..((.((..((.((((.....))))))..)).))..)))).)).)))))))))))(((.((.((((((...)))))...).)).))).)))).. (-32.27 = -32.35 + 0.08)

| Location | 16,857,598 – 16,857,718 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -38.35 |
| Consensus MFE | -32.67 |
| Energy contribution | -32.87 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.02 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979394 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16857598 120 + 27905053 UCCUGAAGGAGUUUGUGAAGCUCAGGCCAACCCUUGAUGGGCAUGUACUUAGGAUCACCAAUUCCUCCAGCGAAAUGACAGUAAAUAUUCGGCUUGUCGAAUAUGACGGACUCGUCAAUU (((((.(((((((.((((..((.((..((.(((.....)))..))..)).))..)))).))))))).))).))..(((((((..((((((((....)))))))).....))).))))... ( -38.90) >DroVir_CAF1 11138 120 + 1 UCCUGAAGGAGUUUGUGAAGCUCAGGCCAACCCUUGAUGGGCAUGUACUUAGGAUCACCAAUUCCUCCAGCGAAAUGACAGUAAAUAUUCGGUUUGUCGAAUAUAACAGAUUCGUCAAUU (((((.(((((((.((((..((.((..((.(((.....)))..))..)).))..)))).))))))).))).))..(((((((..((((((((....)))))))).....))).))))... ( -34.40) >DroPse_CAF1 18942 120 + 1 UCCUGAAGGAGUUUGUGAAGCUCAGUCCAGCCCUUGAUGGGCAUGUACUUAGGAUCACCAAUUCCUCCAGCGAAAUGACAGUAGAUAUUCGGCUUGUCGAAUAUAACGGAUUCGUCAAUU (((((.(((((((.((((..((.(((.((((((.....)))).)).))).))..)))).))))))).))).))..(((((((..((((((((....)))))))).....))).))))... ( -41.50) >DroGri_CAF1 12773 120 + 1 UCCUGAAGGAGUUUGUGAAGCUCAGGCCAGGCCUUGAUUGGCAUGUACUUAGGAUCACCAAUUCCUCCAGCGAAAUGACAGUAAAUAUUCGGCUUGUCGAAUAUCACGGAUUCGUCAAUU (((((.(((((((.((((..((.((..((.(((......))).))..)).))..)))).))))))).))).))..(((((((..((((((((....)))))))).....))).))))... ( -37.60) >DroYak_CAF1 12606 120 + 1 UCCUGAAGGAGUUUGUGAAGCUCAGGCCAACCCUUGAUGGGCAUGUACUUAGGAUCACCAAUUCCUCCAGCGAAAUGACAGUAAAUAUUCGGCUUGUCGAAUAUGACGGAUUCGUCUAUU (((((.(((((((.((((..((.((..((.(((.....)))..))..)).))..)))).))))))).))).))...........((((((((....))))))))((((....)))).... ( -36.20) >DroPer_CAF1 19545 120 + 1 UCCUGAAGGAGUUUGUGAAGCUCAGUCCAGCCCUUGAUGGGCAUGUACUUAGGAUCACCAAUUCCUCCAGCGAAAUGACAGUAGAUAUUCGGCUUGUCGAAUAUAACGGAUUCGUCAAUU (((((.(((((((.((((..((.(((.((((((.....)))).)).))).))..)))).))))))).))).))..(((((((..((((((((....)))))))).....))).))))... ( -41.50) >consensus UCCUGAAGGAGUUUGUGAAGCUCAGGCCAACCCUUGAUGGGCAUGUACUUAGGAUCACCAAUUCCUCCAGCGAAAUGACAGUAAAUAUUCGGCUUGUCGAAUAUAACGGAUUCGUCAAUU (((((.(((((((.((((..((.((..((..(((....)))..))..)).))..)))).))))))).))).))..(((((((..((((((((....)))))))).....))).))))... (-32.67 = -32.87 + 0.20)



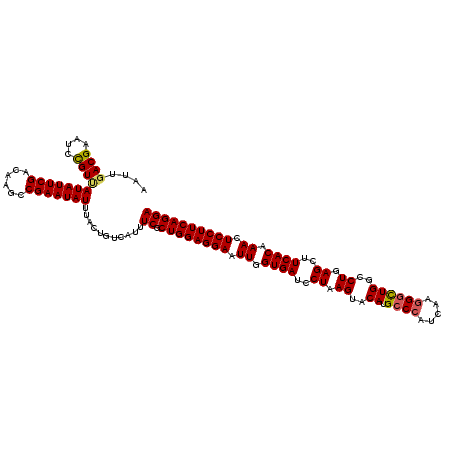
| Location | 16,857,598 – 16,857,718 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -37.77 |
| Consensus MFE | -33.51 |
| Energy contribution | -33.45 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.09 |
| SVM RNA-class probability | 0.998416 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16857598 120 - 27905053 AAUUGACGAGUCCGUCAUAUUCGACAAGCCGAAUAUUUACUGUCAUUUCGCUGGAGGAAUUGGUGAUCCUAAGUACAUGCCCAUCAAGGGUUGGCCUGAGCUUCACAAACUCCUUCAGGA ...(((((....)))))((((((......))))))............((.((((((((.((.((((..((.((..((.((((.....))))))..)).))..)))).)).)))))))))) ( -37.50) >DroVir_CAF1 11138 120 - 1 AAUUGACGAAUCUGUUAUAUUCGACAAACCGAAUAUUUACUGUCAUUUCGCUGGAGGAAUUGGUGAUCCUAAGUACAUGCCCAUCAAGGGUUGGCCUGAGCUUCACAAACUCCUUCAGGA ...(((((.....((.(((((((......)))))))..)))))))..((.((((((((.((.((((..((.((..((.((((.....))))))..)).))..)))).)).)))))))))) ( -36.30) >DroPse_CAF1 18942 120 - 1 AAUUGACGAAUCCGUUAUAUUCGACAAGCCGAAUAUCUACUGUCAUUUCGCUGGAGGAAUUGGUGAUCCUAAGUACAUGCCCAUCAAGGGCUGGACUGAGCUUCACAAACUCCUUCAGGA ...(((((.....((.(((((((......)))))))..)))))))..((.((((((((.((.((((..((.(((.((.((((.....)))))).))).))..)))).)).)))))))))) ( -38.90) >DroGri_CAF1 12773 120 - 1 AAUUGACGAAUCCGUGAUAUUCGACAAGCCGAAUAUUUACUGUCAUUUCGCUGGAGGAAUUGGUGAUCCUAAGUACAUGCCAAUCAAGGCCUGGCCUGAGCUUCACAAACUCCUUCAGGA ...(((((.....((((((((((......)))))).)))))))))..((.((((((((.((.((((..((.((..((.(((......))).))..)).))..)))).)).)))))))))) ( -37.40) >DroYak_CAF1 12606 120 - 1 AAUAGACGAAUCCGUCAUAUUCGACAAGCCGAAUAUUUACUGUCAUUUCGCUGGAGGAAUUGGUGAUCCUAAGUACAUGCCCAUCAAGGGUUGGCCUGAGCUUCACAAACUCCUUCAGGA ....((((....))))(((((((......)))))))...........((.((((((((.((.((((..((.((..((.((((.....))))))..)).))..)))).)).)))))))))) ( -37.60) >DroPer_CAF1 19545 120 - 1 AAUUGACGAAUCCGUUAUAUUCGACAAGCCGAAUAUCUACUGUCAUUUCGCUGGAGGAAUUGGUGAUCCUAAGUACAUGCCCAUCAAGGGCUGGACUGAGCUUCACAAACUCCUUCAGGA ...(((((.....((.(((((((......)))))))..)))))))..((.((((((((.((.((((..((.(((.((.((((.....)))))).))).))..)))).)).)))))))))) ( -38.90) >consensus AAUUGACGAAUCCGUUAUAUUCGACAAGCCGAAUAUUUACUGUCAUUUCGCUGGAGGAAUUGGUGAUCCUAAGUACAUGCCCAUCAAGGGCUGGCCUGAGCUUCACAAACUCCUUCAGGA ....((((....))))(((((((......)))))))...........((.((((((((.((.((((..((.((..((.((((.....))))))..)).))..)))).)).)))))))))) (-33.51 = -33.45 + -0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:21:29 2006