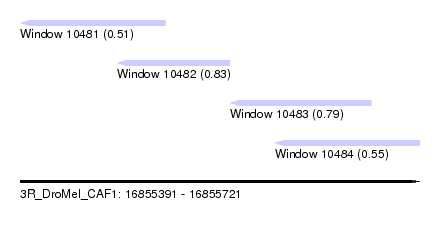
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,855,391 – 16,855,721 |
| Length | 330 |
| Max. P | 0.826645 |

| Location | 16,855,391 – 16,855,511 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.78 |
| Mean single sequence MFE | -31.71 |
| Consensus MFE | -25.82 |
| Energy contribution | -24.71 |
| Covariance contribution | -1.11 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.81 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.513622 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16855391 120 - 27905053 AAGCAGGGCAUUACGUUGCCCGGCGAUAUUCUACUCAUCACCGCCUUUAUUUCCUAUGUCGGCUGUUUCACGAAAGGAUUCCGCAUCGACCUCUUACUCAAGAUGUGGACACCCUUUUUG ((((((((((......)))))((.(((.........))).))(((..(((.....)))..))))))))...((((((..((((((((..............))))))))...)))))).. ( -34.34) >DroVir_CAF1 9007 120 - 1 AAACAGGGCAUUACAUUGCCUGGCGAUAUACUACUCAUUACGGCCUUCAUUUCCUAUGUCGGCUGUUUUACGAAGGGUUUUCGCAUUGAUCUAAUGCAAAAGAUGUGGACACCCUUCCUC .....(((((......)))))..................((((((..(((.....)))..)))))).....(((((((.((((((((..............)))))))).)))))))... ( -36.04) >DroGri_CAF1 10521 120 - 1 AAGCAGGGAAUUACGUUACCCGGCGAUAUACUACUCAUAACCGCCUUCAUUUCCUAUGUCGGCUGUUUCACGAAAGGCUUUCGCAUUGAUCUAUUGCUAAAGAUGUGGACACCCUUUCUG ..((.((((((...))).))).))..............(((.(((..(((.....)))..))).)))....((((((..((((((((..............))))))))...)))))).. ( -26.64) >DroWil_CAF1 3304 120 - 1 AAGCAGGGUAUUACUUUACCCGGCGAUAUACUACUCAUUACCGCCUUCAUUUCCUAUGUUGGCUGUUUCACGAAGGGUUUUCGCAUCGAUCUACUAAUGAAGAUGUGGACACCCUUUCUG ..((.(((((......))))).))...............((.(((..(((.....)))..))).))..((.(((((((.((((((((..((.......)).)))))))).))))))).)) ( -37.70) >DroMoj_CAF1 10554 120 - 1 AAGCAGGGCAUUACCUUACCCGGCGAUAUACUACUCAUUACCGCCUUCAUUUCCUAUGUCGGUUGUUUUACGAAGGGUUUUCGCAUUGAUCUCUUGCAAAAGAUGUGGACACCCUUUCUA (((((((((((..........((((.((..........)).))))..........)))))..))))))...(((((((.((((((((..............)))))))).)))))))... ( -27.29) >DroAna_CAF1 7901 120 - 1 AAGCAGGGAAUUACCUUACCCGGCGACAUUCUACUCAUUACCGCCUUUAUUUCCUAUGUCGGCUGUUUUACGAAAGGAUUCCGCAUCGAUCUUCUACUCAAGAUGUGGACUCCCUUUUUG (((.(((((............((((................))))......((((...(((.........))).)))).((((((((..............)))))))).))))).))). ( -28.23) >consensus AAGCAGGGCAUUACCUUACCCGGCGAUAUACUACUCAUUACCGCCUUCAUUUCCUAUGUCGGCUGUUUCACGAAAGGUUUUCGCAUCGAUCUAUUACUAAAGAUGUGGACACCCUUUCUG ..((.(((..........))).))...............((.(((..(((.....)))..))).)).....((((((..((((((((..............))))))))...)))))).. (-25.82 = -24.71 + -1.11)



| Location | 16,855,471 – 16,855,564 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 77.18 |
| Mean single sequence MFE | -16.55 |
| Consensus MFE | -11.43 |
| Energy contribution | -11.32 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826645 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16855471 93 - 27905053 ACAAUAAAAACUACCA--UACUAAAUGA--CAAUGUU-UAUCCUA--AAUCAGCUUCGUCAAGCAGGGCAUUACGUUGCCCGGCGAUAUUCUACUCAUCA ................--......((((--....(..-((((((.--.....((((....)))).(((((......))))))).))))..)...)))).. ( -17.60) >DroVir_CAF1 9087 91 - 1 CCAAUC---AAUGCCAAAUAAU-AAUCAAAUAAU-CU-UAACUCG---AACAGCUUUGUCAAACAGGGCAUUACAUUGCCUGGCGAUAUACUACUCAUUA ......---..(((((..((((-..........(-(.-......)---)...((((((.....)))))).....))))..)))))............... ( -12.70) >DroSec_CAF1 8379 92 - 1 ACAAGU-GAACUUCCA--UACUAAAUGA--CUAUGUU-UAUUCUA--AAUCAGCUUCGUCAAGCAGGGCAUUACGUUGCCCGGCGAUAUUCUACUCAUCA ...(((-(((..((..--.......(((--(...(((-.((....--.)).)))...)))).((.(((((......))))).))))..))).)))..... ( -19.20) >DroEre_CAF1 8872 92 - 1 AUAAAG-AAACUACCA--UACUAAUUGG--CUAUGUU-UAUCCUA--AAUCAGCUUCGUCAAGCAGGGCAUUACGUUGCCCGGCGACAUUCUACUCAUCA ....((-((....(((--.......)))--...((((-...((..--.....((((....)))).(((((......))))))).))))))))........ ( -18.70) >DroYak_CAF1 10449 92 - 1 GUAAGG-AAACUAUCA--UACCUAAUGG--CUAUGUU-UAUCCUA--AAUCAGCUUCGUCAAGCAGGGCAUCACUUUGCCCGGCGAUAUUCUACUCAUCA ((((((-(......((--((((....))--.))))..-..)))).--.(((.((((....)))).(((((......)))))...)))....)))...... ( -21.80) >DroAna_CAF1 7981 95 - 1 ACAAUAUUAACUAACA--UG-UAAUUAA--UUAUCUUGUAUUCCACCAUAAAGCUUCGUCAAGCAGGGAAUUACCUUACCCGGCGACAUUCUACUCAUUA ................--..-.......--......(((...((........((((....)))).(((..........)))))..)))............ ( -9.30) >consensus ACAAUA_AAACUACCA__UACUAAAUGA__CUAUGUU_UAUCCUA__AAUCAGCUUCGUCAAGCAGGGCAUUACGUUGCCCGGCGAUAUUCUACUCAUCA .......................................................(((((.....(((((......)))))))))).............. (-11.43 = -11.32 + -0.11)



| Location | 16,855,564 – 16,855,681 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.27 |
| Mean single sequence MFE | -45.79 |
| Consensus MFE | -30.87 |
| Energy contribution | -30.57 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.794411 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16855564 117 - 27905053 CAGGAAGCGGAUGCCACCCAAGCCACCAUUGCCCUGGCCAAUCGCCUGGUUGGCGGUCUGGCCAGUGAGAACGUACGUUGGGCCGAAGCGGUCAACAAGUGCGUAUCCGGAG-UA--CAU .......((((((((((....(((.(....((((((((((((((((.....)))))).)))))).....(((....)))))))....).)))......))).)))))))...-..--... ( -44.50) >DroVir_CAF1 9178 118 - 1 CAAGAAGCGGAUGCCACACAGGCAACCAUUGCGCUGGCCAAUCGCCUGGUCGGCGGCCUGGCCAGCGAGAAUGUGCGCUGGGCCGAGGCUGUCAAUAAGUAAGUAGCAUUAGCUA--CUU ......((((.((((.....)))).))(((((((((((((......))))))))(((((((((((((........)))).)))).)))))).))))..))(((((((....))))--))) ( -53.10) >DroGri_CAF1 10701 111 - 1 CAGGAAGCGGAUGCCACACAAGCGACCAUCGCGCUGGCCAAUCGUCUGGUGGGCGGUUUGGCCAGUGAAAACGUGCGCUGGGCUGAAGCAGUCAAUAAGUGAGUAGUC---------AUC ........(..((((((...((((((.....(((((((((((((((.....))))).)))))))))).....)).)))).(((((...))))).....))).)))..)---------... ( -41.50) >DroEre_CAF1 8964 119 - 1 CAGGAAGCGGAUGCUACCCAAGCCACCAUUGCCCUGGCCAAUCGCCUGGUGGGCGGCUUGGCCAGUGAGAACGUACGGUGGGCCGAAGCAGUAAACAAGUGGGUAUUCUAAG-UAUGCAU ........(.((((((((((.......(((((.(((((((((((((.....))))).))))))))..........(((....)))..))))).......)))))......))-))).).. ( -42.34) >DroMoj_CAF1 10730 111 - 1 CAGGAGGCGGAUGCCACACAGGCAACCAUUGCGCUGGCCAAUCGUCUGGUCGGCGGUCUGGCCAGCGAGAACGUACGCUGGGCUGAGGCAGUCAAUAAGUGAGUAGUA---------CUC ...(((..((.((((.....)))).))(((((((((((((((((((.....)))))).)))))))).........((((.(((((...)))))....)))).))))).---------))) ( -46.10) >DroAna_CAF1 8076 114 - 1 CAGGAGGCGGACGCCACUCAAGCAACCAUCGCGCUGGCCAAUCGCCUGGUGGGCGGCCUGGCCAGUGAAAAUGUCCGCUGGGCGGAAGCAGUCAACAAGUGAGAUUCCAAAU-UA----- ..((((((((((((.......))........(((((((((.(((((.....)))))..))))))))).....)))))))..((....))................)))....-..----- ( -47.20) >consensus CAGGAAGCGGAUGCCACACAAGCAACCAUUGCGCUGGCCAAUCGCCUGGUGGGCGGCCUGGCCAGUGAGAACGUACGCUGGGCCGAAGCAGUCAACAAGUGAGUAGCC__AG_UA__CUU ......((....(((.....(((.((.....(((((((((..((((.....))))...))))))))).....))..))).)))....))............................... (-30.87 = -30.57 + -0.30)


| Location | 16,855,601 – 16,855,721 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.50 |
| Mean single sequence MFE | -45.60 |
| Consensus MFE | -32.61 |
| Energy contribution | -32.12 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.551224 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16855601 120 - 27905053 GGACUUUGAGAAGGCCACUGCAGAUAAGUUGCGGUGCCAACAGGAAGCGGAUGCCACCCAAGCCACCAUUGCCCUGGCCAAUCGCCUGGUUGGCGGUCUGGCCAGUGAGAACGUACGUUG ((.(..((....(((.(((((((.....))))))))))....((....((......))....))..))..)))(((((((((((((.....)))))).)))))))....(((....))). ( -41.70) >DroVir_CAF1 9216 120 - 1 AGACUUUGAGCAGGCCAUUGCGGACAAAUUACGUUGUCAACAAGAAGCGGAUGCCACACAGGCAACCAUUGCGCUGGCCAAUCGCCUGGUCGGCGGCCUGGCCAGCGAGAAUGUGCGCUG ........(((......(((..(((((......)))))..)))...((((.((((.....)))).))....(((((((((.(((((.....)))))..))))))))).......))))). ( -48.20) >DroGri_CAF1 10732 120 - 1 GGACUUUGAGACGGCCAUUGCCGAUAAAUUGCGUUGUCAACAGGAAGCGGAUGCCACACAAGCGACCAUCGCGCUGGCCAAUCGUCUGGUGGGCGGUUUGGCCAGUGAAAACGUGCGCUG .(((..((...((((....))))........))..))).......((((.(((........(((.....)))((((((((((((((.....))))).))))))))).....))).)))). ( -42.50) >DroWil_CAF1 3516 120 - 1 CGAUUUUGAGAAAGCCACAGCAGAUAAAUUACGCUGCCAACAGGAAGCGGAUGCCACUCAGGCCACUAUAGCAUUGGCUAACCGCCUAGUUGGUGGUUUGGCCAGUGAAAAUGUACGCUG .................((((..........((((.((....)).))))...(((.....))).((.....(((((((((((((((.....))))).)))))))))).....))..)))) ( -40.50) >DroMoj_CAF1 10761 120 - 1 GGACUUUGAAAAAGCGAUUGCGGAUAAGCUACGUUGUCAGCAGGAGGCGGAUGCCACACAGGCAACCAUUGCGCUGGCCAAUCGUCUGGUCGGCGGUCUGGCCAGCGAGAACGUACGCUG ............((((...((......))((((((.((.((((.....((.((((.....)))).)).))))((((((((((((((.....)))))).)))))))))).)))))))))). ( -51.30) >DroAna_CAF1 8110 120 - 1 GGAUUUCGAGAAGGCUACAGCCGAUAAACUGCGUUGUCAGCAGGAGGCGGACGCCACUCAAGCAACCAUCGCGCUGGCCAAUCGCCUGGUGGGCGGCCUGGCCAGUGAAAAUGUCCGCUG ............(((....)))......((((.......))))..(((((((((.......))........(((((((((.(((((.....)))))..))))))))).....))))))). ( -49.40) >consensus GGACUUUGAGAAGGCCACUGCAGAUAAAUUACGUUGUCAACAGGAAGCGGAUGCCACACAAGCAACCAUUGCGCUGGCCAAUCGCCUGGUCGGCGGUCUGGCCAGUGAAAACGUACGCUG ....((((.....((....)).(((((......)))))..)))).((((.((((.......))........(((((((((((((((.....)))))).))))))))).....)).)))). (-32.61 = -32.12 + -0.49)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:21:24 2006