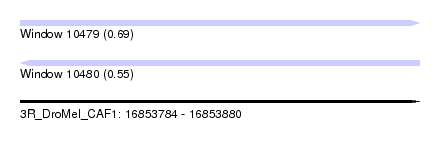
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,853,784 – 16,853,880 |
| Length | 96 |
| Max. P | 0.691922 |

| Location | 16,853,784 – 16,853,880 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 80.45 |
| Mean single sequence MFE | -25.58 |
| Consensus MFE | -15.09 |
| Energy contribution | -14.73 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.691922 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
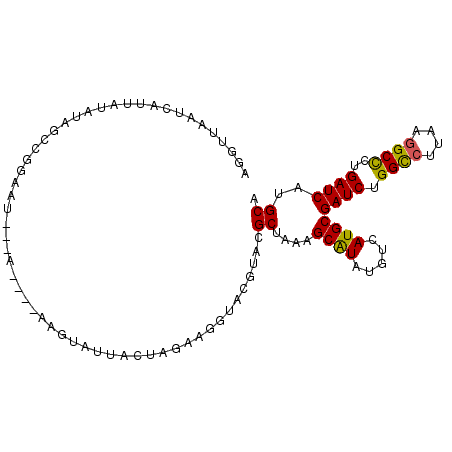
>3R_DroMel_CAF1 16853784 96 + 27905053 AGGUUAAUCAUUAUAUAGCCGGAAU---A----AAGUAUUACUAGAAGGUACGUACGCUAAAGCGUAUGUCAUGCGAUCUGGCCUUAAGGCCCUGAUCAUGCA .(((((.........))))).....---.----..((((.....(...(.((((((((....)))))))))...)((((.((((....))))..)))))))). ( -29.10) >DroSec_CAF1 6693 96 + 1 AGGUUAAUUAUUGUAUAGCCGGAAU---A----AAGUAUUACUAGAAGGUACGUACGCUAAAGCGUAUGUCAUGCGAUCUGGCCUUAAGGCCCUGAUCAUGCA .(((((.........))))).....---.----..((((.....(...(.((((((((....)))))))))...)((((.((((....))))..)))))))). ( -29.10) >DroSim_CAF1 6687 96 + 1 AGAUUAAUCAUUAUAUAGCCGGAAU---A----AAGUAUUACUAGAAGGUACGUACGCUAAAGCGUAUGUCAUGCGAUCUGGCCUUAAGGCCCUGAUCAUGCA .................((......---.----..((((((((....))))(((((((....)))))))..))))((((.((((....))))..))))..)). ( -26.00) >DroEre_CAF1 7183 97 + 1 AAGAUAAUCUUUAUAUAUCCGGUAG------ACAAAUGUUACUAGAAUCUACGUACGCUAAAGCAUAAGUCAUGCGAUCAGGCCUUAAGGCCCUGAUCAUGCA .................((..((.(------((....)))))..))..........((....((((.....))))(((((((((....)).)))))))..)). ( -23.20) >DroYak_CAF1 8757 100 + 1 AAGUUAAUUAUCAUAUAUCCGGAAU---GGAAUAUAUAUUCCUAGAGUUUACGUACGCUAAAGCAUAUGUCAUGCGAUCAGGUCUUAAGGCCCUGAUCAUGCA ....................(((((---(.......))))))....((..(((((.((....)).)))))...))(((((((((....)).)))))))..... ( -23.00) >DroPer_CAF1 15821 97 + 1 GGAUUACACAUUAUUCGUACUUAAUUUCG----GAGUA--UCUGGCGGAUACUUACGCUAAAGCAUAGGUCAUGCGAUCGGGCCUUAAAGCUCUGAUCAUGCA ((((........)))).............----(((((--((.....)))))))..((....((((.....))))((((((((......)).))))))..)). ( -23.10) >consensus AGGUUAAUCAUUAUAUAGCCGGAAU___A____AAGUAUUACUAGAAGGUACGUACGCUAAAGCAUAUGUCAUGCGAUCUGGCCUUAAGGCCCUGAUCAUGCA ........................................................((....((((.....))))((((.((((....))))..))))..)). (-15.09 = -14.73 + -0.36)

| Location | 16,853,784 – 16,853,880 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 80.45 |
| Mean single sequence MFE | -21.84 |
| Consensus MFE | -15.95 |
| Energy contribution | -15.90 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.548607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
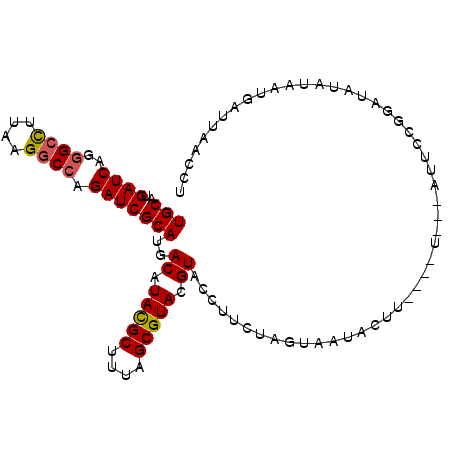
>3R_DroMel_CAF1 16853784 96 - 27905053 UGCAUGAUCAGGGCCUUAAGGCCAGAUCGCAUGACAUACGCUUUAGCGUACGUACCUUCUAGUAAUACUU----U---AUUCCGGCUAUAUAAUGAUUAACCU ..(((((((..((((....)))).))))((...((.(((((....))))).)).......(((((....)----)---)))...))......)))........ ( -21.10) >DroSec_CAF1 6693 96 - 1 UGCAUGAUCAGGGCCUUAAGGCCAGAUCGCAUGACAUACGCUUUAGCGUACGUACCUUCUAGUAAUACUU----U---AUUCCGGCUAUACAAUAAUUAACCU .((..((((..((((....)))).)))).....((.(((((....))))).)).................----.---......))................. ( -20.90) >DroSim_CAF1 6687 96 - 1 UGCAUGAUCAGGGCCUUAAGGCCAGAUCGCAUGACAUACGCUUUAGCGUACGUACCUUCUAGUAAUACUU----U---AUUCCGGCUAUAUAAUGAUUAAUCU ..(((((((..((((....)))).))))((...((.(((((....))))).)).......(((((....)----)---)))...))......)))........ ( -21.10) >DroEre_CAF1 7183 97 - 1 UGCAUGAUCAGGGCCUUAAGGCCUGAUCGCAUGACUUAUGCUUUAGCGUACGUAGAUUCUAGUAACAUUUGU------CUACCGGAUAUAUAAAGAUUAUCUU .((..(((((((.((....)))))))))(((((...)))))....))....((((((.............))------)))).(((((.........))))). ( -25.42) >DroYak_CAF1 8757 100 - 1 UGCAUGAUCAGGGCCUUAAGACCUGAUCGCAUGACAUAUGCUUUAGCGUACGUAAACUCUAGGAAUAUAUAUUCC---AUUCCGGAUAUAUGAUAAUUAACUU (((..(((((((.........))))))))))..((.(((((....))))).))............(((((((((.---.....)))))))))........... ( -23.30) >DroPer_CAF1 15821 97 - 1 UGCAUGAUCAGAGCUUUAAGGCCCGAUCGCAUGACCUAUGCUUUAGCGUAAGUAUCCGCCAGA--UACUC----CGAAAUUAAGUACGAAUAAUGUGUAAUCC (((..((((.(.(((....)))).))))((((......(((((...((..((((((.....))--)))).----)).....)))))......))))))).... ( -19.20) >consensus UGCAUGAUCAGGGCCUUAAGGCCAGAUCGCAUGACAUACGCUUUAGCGUACGUACCUUCUAGUAAUACUU____U___AUUCCGGAUAUAUAAUGAUUAACCU (((..((((..((((....)))).)))))))..((.(((((....))))).)).................................................. (-15.95 = -15.90 + -0.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:21:20 2006