| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,381,328 – 2,381,436 |
| Length | 108 |
| Max. P | 0.779810 |

| Location | 2,381,328 – 2,381,436 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 84.50 |
| Mean single sequence MFE | -30.78 |
| Consensus MFE | -21.21 |
| Energy contribution | -21.30 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.596669 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2381328 108 + 27905053 UAGCUUUUUUGGCAAGCAUCCUGCUGAAUG----GAGGAA----GUGGUGCCACUUU-UUAUGCAUUCGACUGUGCAUAAGUAUGUAUAUACUUCUAUAUAUUUUUGGCCGGCCAAA .......((((((..((.(((........)----))((((----(((((((.....(-((((((((......)))))))))...)))).)))))))...........))..)))))) ( -31.10) >DroSec_CAF1 93942 108 + 1 UAGCUUUUUUGGCAAGCAUCCUGGUGAAUA----GAGGAA----CUGGUGCCACUUU-UUAUGCAUUCGACUGUGCAUAAGUAUGUAUAUACUUCUAUAUAUUUUUCGCCGGCCAAA .......((((((..((.....((((.(((----((((..----...((((.....(-((((((((......)))))))))...))))...))))))).))))....))..)))))) ( -28.40) >DroSim_CAF1 86632 108 + 1 UAGCUUUUUUGGCAAGCAUCCUGGUGAAUG----GAGGAA----CUGGUGCCACUUU-UUAUGCAUUCGACUGUGCAUAAGUAUGUAUAUACUUCUAUAUAUUUUUCGCCGGCCAAA .......((((((..((.....((((.(((----((((..----...((((.....(-((((((((......)))))))))...))))...))))))).))))....))..)))))) ( -28.10) >DroEre_CAF1 88148 103 + 1 -----UUUUUGGCAAGCAUCCUGCUGAAUG----GAGGAG----CUGGUGCCACUUU-UUAUGCAUUCGACUGUGCAUAAGUACGUAUAUACUUCUAUAUAUUUUUGGCCGGCCAAA -----..((((((.(((.((((.(.....)----.)))))----)).(.((((...(-((((((((......)))))))))...(((((((....)))))))...))))).)))))) ( -33.10) >DroYak_CAF1 82347 108 + 1 UAGCUUUUUUGGCAAGCAUCCUGCUGAAUG----GAGGAA----CUGGUGCCACUUU-UUAUGCAUUCGACUGUGCAUAAGUACGUAUAUAUUUCUAUAUAUUUUUGGCCGGCCAAA .......((((((.....((((.(.....)----.)))).----...(.((((...(-((((((((......)))))))))...(((((((....)))))))...))))).)))))) ( -29.50) >DroAna_CAF1 86569 106 + 1 UUUUAUUUUUGGCAAGCAUCCUUCUGAACAGCCAGAGGAGCUAGCUAGUGCUACUCUCCUAUGCAUCAGAGUGUGCAUAAAUACAUAUAUAUUUG-----------UGCCGGCCAAA .......((((((..((((...(((((...((...(((((.((((....))))..)))))..)).)))))))))(((((((((......))))))-----------)))..)))))) ( -34.50) >consensus UAGCUUUUUUGGCAAGCAUCCUGCUGAAUG____GAGGAA____CUGGUGCCACUUU_UUAUGCAUUCGACUGUGCAUAAGUACGUAUAUACUUCUAUAUAUUUUUGGCCGGCCAAA .......((((((..((.((((.............))))........((((.......((((((((......)))))))))))).......................))..)))))) (-21.21 = -21.30 + 0.08)
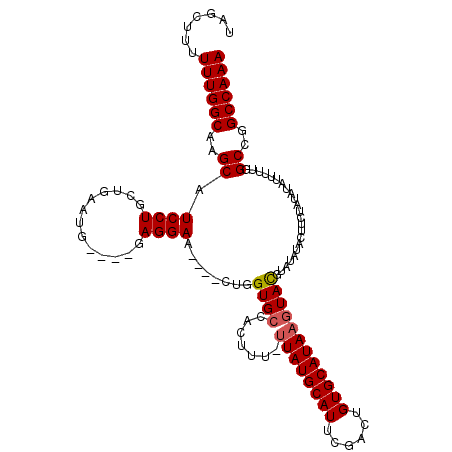

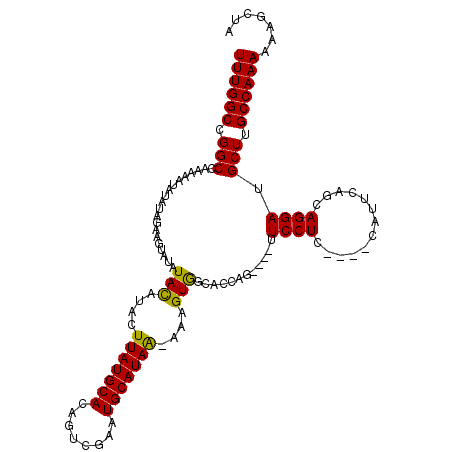
| Location | 2,381,328 – 2,381,436 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 84.50 |
| Mean single sequence MFE | -26.57 |
| Consensus MFE | -20.85 |
| Energy contribution | -20.44 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2381328 108 - 27905053 UUUGGCCGGCCAAAAAUAUAUAGAAGUAUAUACAUACUUAUGCACAGUCGAAUGCAUAA-AAAGUGGCACCAC----UUCCUC----CAUUCAGCAGGAUGCUUGCCAAAAAAGCUA ((((((.(((...............(((((........)))))...((.(((((.....-.(((((....)))----))....----))))).)).....))).))))))....... ( -25.00) >DroSec_CAF1 93942 108 - 1 UUUGGCCGGCGAAAAAUAUAUAGAAGUAUAUACAUACUUAUGCACAGUCGAAUGCAUAA-AAAGUGGCACCAG----UUCCUC----UAUUCACCAGGAUGCUUGCCAAAAAAGCUA ((((((.((((....((((((....)))))).(((..(((((((........)))))))-...))).......----.((((.----........)))))))).))))))....... ( -22.70) >DroSim_CAF1 86632 108 - 1 UUUGGCCGGCGAAAAAUAUAUAGAAGUAUAUACAUACUUAUGCACAGUCGAAUGCAUAA-AAAGUGGCACCAG----UUCCUC----CAUUCACCAGGAUGCUUGCCAAAAAAGCUA ((((((.((((((..((((((....)))))).(((..(((((((........)))))))-...))).......----))).((----(........))).))).))))))....... ( -23.10) >DroEre_CAF1 88148 103 - 1 UUUGGCCGGCCAAAAAUAUAUAGAAGUAUAUACGUACUUAUGCACAGUCGAAUGCAUAA-AAAGUGGCACCAG----CUCCUC----CAUUCAGCAGGAUGCUUGCCAAAAA----- ((((((..((((...((((((....))))))......(((((((........)))))))-....))))...((----(((((.----(.....).)))).))).))))))..----- ( -27.20) >DroYak_CAF1 82347 108 - 1 UUUGGCCGGCCAAAAAUAUAUAGAAAUAUAUACGUACUUAUGCACAGUCGAAUGCAUAA-AAAGUGGCACCAG----UUCCUC----CAUUCAGCAGGAUGCUUGCCAAAAAAGCUA ((((((..((((....((((((....)))))).....(((((((........)))))))-....))))...((----(((((.----(.....).)))).))).))))))....... ( -27.10) >DroAna_CAF1 86569 106 - 1 UUUGGCCGGCA-----------CAAAUAUAUAUGUAUUUAUGCACACUCUGAUGCAUAGGAGAGUAGCACUAGCUAGCUCCUCUGGCUGUUCAGAAGGAUGCUUGCCAAAAAUAAAA ((((((.((((-----------.((((((....)))))).....(..(((((.((..(((((..((((....)))).)))))...))...)))))..).)))).))))))....... ( -34.30) >consensus UUUGGCCGGCCAAAAAUAUAUAGAAGUAUAUACAUACUUAUGCACAGUCGAAUGCAUAA_AAAGUGGCACCAG____UUCCUC____CAUUCAGCAGGAUGCUUGCCAAAAAAGCUA ((((((.(((....................(((....(((((((........)))))))....)))............((((.............)))).))).))))))....... (-20.85 = -20.44 + -0.42)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:10 2006