| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,378,527 – 2,378,619 |
| Length | 92 |
| Max. P | 0.922683 |

| Location | 2,378,527 – 2,378,619 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 84.03 |
| Mean single sequence MFE | -15.18 |
| Consensus MFE | -14.87 |
| Energy contribution | -14.87 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
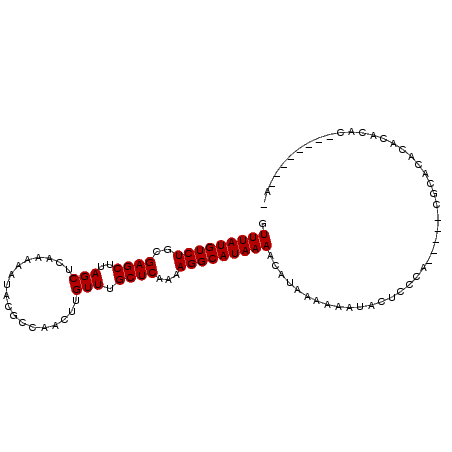
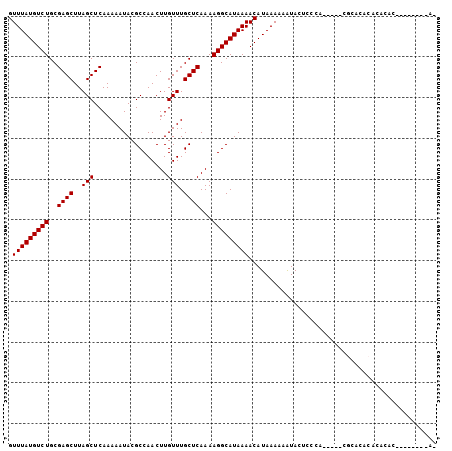
>3R_DroMel_CAF1 2378527 92 - 27905053 GUUUAUGUCUGCGAGCUUAGCUCAAAAAUACGCCAACUUGUUUGCUCAAAAGGCAUAAAACAUAAAAAAUACUACCA-----CGAACACACACAC--------AC .(((((((((..((((..(((..................))).))))...)))))))))..................-----.............--------.. ( -14.87) >DroSec_CAF1 91130 92 - 1 GUUUAUGUCUGCGAGCUUAGCUCAAAAAUACGCCAACUUGUUUGCUCAAAAGGCAUAAAACAUAAAAAAUAC----A-----CGCAC----ACACACACACAGAU ((.(.(((.(((((((...))))........(((...(((......)))..)))..................----.-----.))).----))).).))...... ( -15.00) >DroSim_CAF1 83816 100 - 1 GUUUAUGUCUGCGAGCUUAGCUCAAAAAUACGCCAACUUGUUUGCUCAAAAGGCAUAAAACAUAAAAAAUACUCCCA-----CGCACACACACACACACAGAGAU ((.(.(((.(((((((...))))........(((...(((......)))..))).......................-----.))).))).).)).......... ( -15.00) >DroEre_CAF1 85618 87 - 1 GUUUAUGUCUGCGAGCUUAGCUCAAAAAUACGCCAACUUGUUUGCUCAAAAGGCAUAAAACAUAAAAAAUACUCCCA-----CGCACACACA------------- .(((((((((..((((..(((..................))).))))...)))))))))..................-----..........------------- ( -14.87) >DroYak_CAF1 79591 87 - 1 GUUUAUGUCUGCGAGCUUAGCUCAAAAAUACGCCAACUUGUUUGCUCAAAAGGCAUAAAACAUAAAAAAUACUCACA-----CGCACACAAA------------- .....(((.(((((((...))))........(((...(((......)))..))).......................-----.))).)))..------------- ( -15.10) >DroAna_CAF1 80819 103 - 1 GUUUAUGUCUGCGAGCUUAGCUCAAAAAUACGCCAACUUGUUUGCUCAAAAGGCAUAAAACAUAAAAAGUACCCCCCGUUCCCCAACACAACAGGACACAU--AG .(((((((((..((((..(((..................))).))))...)))))))))..................(((((...........))).))..--.. ( -16.27) >consensus GUUUAUGUCUGCGAGCUUAGCUCAAAAAUACGCCAACUUGUUUGCUCAAAAGGCAUAAAACAUAAAAAAUACUCCCA_____CGCACACACACAC________A_ .(((((((((..((((..(((..................))).))))...))))))))).............................................. (-14.87 = -14.87 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:08 2006