
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,778,012 – 16,778,146 |
| Length | 134 |
| Max. P | 0.998960 |

| Location | 16,778,012 – 16,778,114 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.13 |
| Mean single sequence MFE | -28.94 |
| Consensus MFE | -14.42 |
| Energy contribution | -14.58 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.50 |
| SVM decision value | 3.30 |
| SVM RNA-class probability | 0.998960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16778012 102 + 27905053 GGAAAAUGUUUUCCAAAUGAUGCAAACAAUUGAAAGAUUGUUCGAAUUCUCGCAGAAGAUACAUACAGUUGU-------------AUAUGUAGCUCAAUUGUGUGUG-----AAUCUGUG (((((....)))))....((..(.(((((((....))))))).)...)).((((((...((((((((((((.-------------..........))))))))))))-----..)))))) ( -29.70) >DroSec_CAF1 10381 106 + 1 GGAAAAUGUUUUCCAAAUGAUGCAGACAAUUGAAAGAUUGUUCGAAUUCUCGCAGAAGAUACACACAGUUGUACGU---------AUAUGUAGCUCAAUUGUGUGUG-----AAUCUGUG (((((....)))))....((..(..((((((....))))))..)...)).((((((...((((((((((((((((.---------...))))...))))))))))))-----..)))))) ( -33.20) >DroSim_CAF1 12065 110 + 1 GGAAAAUGUUUUCCAAAUGAUGCAGACAAUUGAAAGAUUGUUCGAAUUCUCGCAGAAGAUACACACAGUUGUACGUAU-----GUAUAUGUAGCUCAAUUGUGUUUG-----AAUCUGUG (((((....)))))....((..(..((((((....))))))..)...)).((((((...((.(((((((((((((((.-----...))))))...))))))))).))-----..)))))) ( -29.90) >DroEre_CAF1 6218 101 + 1 GGAAAAUGUUUUCCAAACGAUGCAGACAGUUGAAAGAUUGUUCGAAUUCUUGCAGGAGAUACACACAGUUGU-------------AUA-GUAGCUCAAUUGUGUGUG-----AAUCUGUG (((((....)))))....((..(..((((((....))))))..)...))..(((((...((((((((((((.-------------...-......))))))))))))-----..))))). ( -30.60) >DroWil_CAF1 4152 109 + 1 AUAAAAUGUAUCUU--AUAGCUCGGGAAACUAGACGAUGGUAUGCAU---------AGGUAUAUAUGUAUGCACAUAUAUUGUUUAAAUGUAACUCAAUUGUGCAUGGAGUCAAUUCGUG ..............--...((((((....))((((((((((((((((---------(......))))))))).....))))))))..(((((.(......)))))).))))......... ( -21.30) >consensus GGAAAAUGUUUUCCAAAUGAUGCAGACAAUUGAAAGAUUGUUCGAAUUCUCGCAGAAGAUACACACAGUUGUAC_U_________AUAUGUAGCUCAAUUGUGUGUG_____AAUCUGUG (((((....))))).......(((((...(((((......)))))..............((((((((((((........................)))))))))))).......))))). (-14.42 = -14.58 + 0.16)

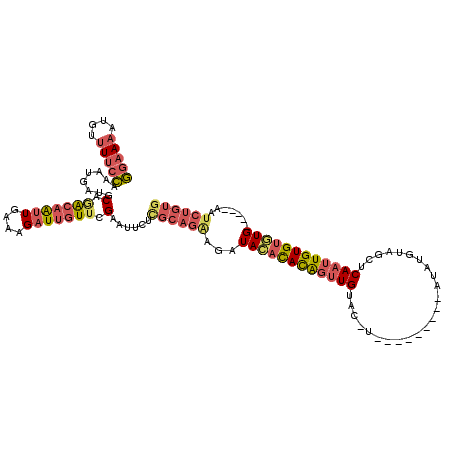

| Location | 16,778,012 – 16,778,114 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.13 |
| Mean single sequence MFE | -18.88 |
| Consensus MFE | -6.32 |
| Energy contribution | -7.08 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.33 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.580185 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
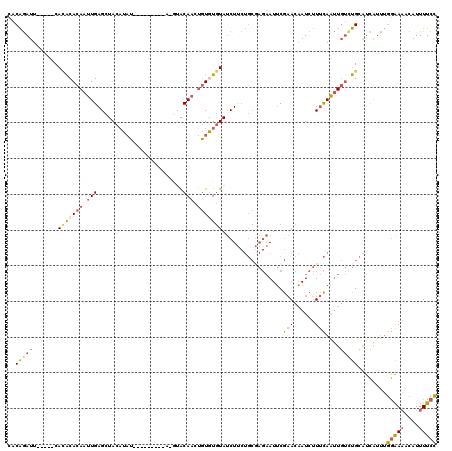
>3R_DroMel_CAF1 16778012 102 - 27905053 CACAGAUU-----CACACACAAUUGAGCUACAUAU-------------ACAACUGUAUGUAUCUUCUGCGAGAAUUCGAACAAUCUUUCAAUUGUUUGCAUCAUUUGGAAAACAUUUUCC ....(((.-----..((.(((((((((.(((((((-------------(....))))))))((((....)))).............))))))))).)).)))....(((((....))))) ( -18.60) >DroSec_CAF1 10381 106 - 1 CACAGAUU-----CACACACAAUUGAGCUACAUAU---------ACGUACAACUGUGUGUAUCUUCUGCGAGAAUUCGAACAAUCUUUCAAUUGUCUGCAUCAUUUGGAAAACAUUUUCC ...((((.-----.(((((((.(((.((.......---------..)).))).)))))))))))..(((((.((((.(((......))))))).)).)))......(((((....))))) ( -20.90) >DroSim_CAF1 12065 110 - 1 CACAGAUU-----CAAACACAAUUGAGCUACAUAUAC-----AUACGUACAACUGUGUGUAUCUUCUGCGAGAAUUCGAACAAUCUUUCAAUUGUCUGCAUCAUUUGGAAAACAUUUUCC ....(((.-----((...((((((((((.....((((-----(((((......))))))))).....))((((..........)))))))))))).)).)))....(((((....))))) ( -22.00) >DroEre_CAF1 6218 101 - 1 CACAGAUU-----CACACACAAUUGAGCUAC-UAU-------------ACAACUGUGUGUAUCUCCUGCAAGAAUUCGAACAAUCUUUCAACUGUCUGCAUCGUUUGGAAAACAUUUUCC ...((((.-----.(((((((.(((......-...-------------.))).)))))))))))..((((.((....(((......))).....))))))......(((((....))))) ( -16.80) >DroWil_CAF1 4152 109 - 1 CACGAAUUGACUCCAUGCACAAUUGAGUUACAUUUAAACAAUAUAUGUGCAUACAUAUAUACCU---------AUGCAUACCAUCGUCUAGUUUCCCGAGCUAU--AAGAUACAUUUUAU .((((..((((((.((.....)).))))))...............((((((((..........)---------)))))))...)))).((((((...)))))).--.............. ( -16.10) >consensus CACAGAUU_____CACACACAAUUGAGCUACAUAU_________A_GUACAACUGUGUGUAUCUUCUGCGAGAAUUCGAACAAUCUUUCAAUUGUCUGCAUCAUUUGGAAAACAUUUUCC ..(((((.......(((((((.(((........................))).))))))).................(((......)))....)))))........(((((....))))) ( -6.32 = -7.08 + 0.76)


| Location | 16,778,052 – 16,778,146 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 92.79 |
| Mean single sequence MFE | -25.43 |
| Consensus MFE | -23.33 |
| Energy contribution | -23.21 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963282 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16778052 94 + 27905053 UUCGAAUUCUCGCAGAAGAUACAUACAGUUGU--------AUAUGUAGCUCAAUUGUGUGUGAAUCUGUGCCUGUAUCUUUUGAUUCGCACUUUGCUUGUUG ...........((((((((((((((((((((.--------..........)))))))))))..))))((((....(((....)))..)))))))))...... ( -23.70) >DroSec_CAF1 10421 98 + 1 UUCGAAUUCUCGCAGAAGAUACACACAGUUGUACGU----AUAUGUAGCUCAAUUGUGUGUGAAUCUGUGCCUGUAUCUUUUGAUUCGCACUUUGCUUGUUG ...........((((((((((((((((((((((((.----...))))...)))))))))))..))))((((....(((....)))..)))))))))...... ( -26.60) >DroSim_CAF1 12105 102 + 1 UUCGAAUUCUCGCAGAAGAUACACACAGUUGUACGUAUGUAUAUGUAGCUCAAUUGUGUUUGAAUCUGUGCCUGUAUCUUUUGAUUCGCACUUUGCUUGUUG ...((((.....((((((((((((((((...((((((....))))))..((((......))))..)))))..)))))))))))))))((.....))...... ( -24.70) >DroEre_CAF1 6258 93 + 1 UUCGAAUUCUUGCAGGAGAUACACACAGUUGU--------AUA-GUAGCUCAAUUGUGUGUGAAUCUGUGCCUGUAUCUUUUGAUUCGCACUUUGCUUGUUG .(((((....(((((((((((((((((((((.--------...-......)))))))))))..))))...))))))...)))))...((.....))...... ( -26.70) >consensus UUCGAAUUCUCGCAGAAGAUACACACAGUUGU________AUAUGUAGCUCAAUUGUGUGUGAAUCUGUGCCUGUAUCUUUUGAUUCGCACUUUGCUUGUUG ...........((((((((((((((((((((...................)))))))))))..))))((((....(((....)))..)))))))))...... (-23.33 = -23.21 + -0.12)

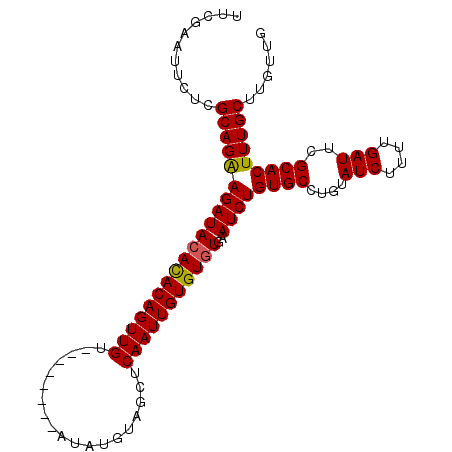

| Location | 16,778,052 – 16,778,146 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 92.79 |
| Mean single sequence MFE | -19.40 |
| Consensus MFE | -15.66 |
| Energy contribution | -16.16 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.695240 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
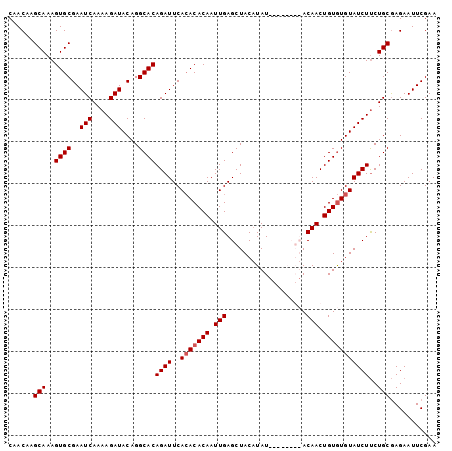
>3R_DroMel_CAF1 16778052 94 - 27905053 CAACAAGCAAAGUGCGAAUCAAAAGAUACAGGCACAGAUUCACACACAAUUGAGCUACAUAU--------ACAACUGUAUGUAUCUUCUGCGAGAAUUCGAA ......(((..(((.(((((................))))).)))......(((.(((((((--------(....)))))))).))).)))........... ( -18.19) >DroSec_CAF1 10421 98 - 1 CAACAAGCAAAGUGCGAAUCAAAAGAUACAGGCACAGAUUCACACACAAUUGAGCUACAUAU----ACGUACAACUGUGUGUAUCUUCUGCGAGAAUUCGAA ......(((..((((..(((....)))....))))((((..(((((((.(((.((.......----..)).))).)))))))))))..)))........... ( -20.80) >DroSim_CAF1 12105 102 - 1 CAACAAGCAAAGUGCGAAUCAAAAGAUACAGGCACAGAUUCAAACACAAUUGAGCUACAUAUACAUACGUACAACUGUGUGUAUCUUCUGCGAGAAUUCGAA ..............(((((((.(((((((..((((((.(((((......))))).......(((....)))...))))))))))))).)).....))))).. ( -19.60) >DroEre_CAF1 6258 93 - 1 CAACAAGCAAAGUGCGAAUCAAAAGAUACAGGCACAGAUUCACACACAAUUGAGCUAC-UAU--------ACAACUGUGUGUAUCUCCUGCAAGAAUUCGAA ......(((..((((..(((....)))....))))((((..(((((((.(((......-...--------.))).)))))))))))..)))........... ( -19.00) >consensus CAACAAGCAAAGUGCGAAUCAAAAGAUACAGGCACAGAUUCACACACAAUUGAGCUACAUAU________ACAACUGUGUGUAUCUUCUGCGAGAAUUCGAA ......(((..((((..(((....)))....))))((((..(((((((.(((...................))).)))))))))))..)))........... (-15.66 = -16.16 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:20:41 2006