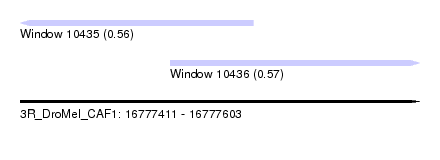
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,777,411 – 16,777,603 |
| Length | 192 |
| Max. P | 0.569518 |

| Location | 16,777,411 – 16,777,523 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 90.03 |
| Mean single sequence MFE | -32.22 |
| Consensus MFE | -24.59 |
| Energy contribution | -26.22 |
| Covariance contribution | 1.63 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.556909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16777411 112 - 27905053 CCAUUGUUGAGCUCCGGCUCAGCUACUUCAGGUAGCUGGACAAUAAACUACCCCUUCGGAACUAAGCCACCCCUGAAGGGUAAUGAUCGAUGGACCUUGAGAGACCUACAAG ((((((..((((....))))((((((.....))))))..............((((((((.............)))))))).......))))))..((((((....)).)))) ( -32.72) >DroSec_CAF1 9811 104 - 1 CCAUUGUUGAGCCCCGGCUCAGCUACUUCAGGUA----GACAAUAAACCACCCCUUCGGAACUGAGCCACCCCUGAAGGGUAUUGAUCGAUGGACCUUGAGAGA----CAAG .....(((((((....)))))))..(((.((((.----(((((((......((((((((.............))))))))))))).)).....)))).)))...----.... ( -31.52) >DroSim_CAF1 11464 112 - 1 CCAUUGUUGAGCCCCGGCUCAGCUACUUCAGGUAGCUGGACAAUAAACCACCCCUUCGGAACUAAGCCACCCCUGAAGCGAAUUGAUCGAUGGACCUUGAGAGACCUACAAG ((((((((((((....)))))))..(((((((..(((((........))....(....).....)))....)))))))..........)))))..((((((....)).)))) ( -31.70) >DroEre_CAF1 5630 111 - 1 CCAUUGUUGAGCUCCGGCUCAGCUACUUCAGGUAGCUGGACAAUAAACCACC-CUUCUAACUUAAGCCACCCCUGAAGGGUAUUGAUCGGUAGACCUUGAGAAACCUACAAG .....(((((((....)))))))..(((.((((.(((((.((((.....(((-((((.................))))))))))).)))))..)))).)))........... ( -32.93) >consensus CCAUUGUUGAGCCCCGGCUCAGCUACUUCAGGUAGCUGGACAAUAAACCACCCCUUCGGAACUAAGCCACCCCUGAAGGGUAUUGAUCGAUGGACCUUGAGAGACCUACAAG .....(((((((....)))))))..(((.((((...(.(((((((......((((((((.............))))))))))))).)).)...)))).)))........... (-24.59 = -26.22 + 1.63)


| Location | 16,777,483 – 16,777,603 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.47 |
| Mean single sequence MFE | -39.35 |
| Consensus MFE | -32.74 |
| Energy contribution | -33.43 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16777483 120 + 27905053 UCCAGCUACCUGAAGUAGCUGAGCCGGAGCUCAACAAUGGCUCAAUUGUUCCUGUUCGCCACCGGUUCAAACGAGUUUCGGGCGGUAAGACGAGAUUUGUGAAUCUCCAGAACUCUGAAC ...((((((.....))))))(((((((.((..((((..(((......)))..)))).))..)))))))..((((((((((..(.....).)))))))))).......(((....)))... ( -36.70) >DroSec_CAF1 9879 116 + 1 UC----UACCUGAAGUAGCUGAGCCGGGGCUCAACAAUGGCUCAAUUGUUCCUGUUCGCCACUGGUUCAAACGAGUUUCGGGCGGUAAGACGAGAUUCGUGGAUCUCCAGAACUCUGAAC .(----..(((((((....(((((((((((..((((..(((......)))..)))).))).))))))))......)))))))..)......((((((....))))))(((....)))... ( -37.10) >DroSim_CAF1 11536 120 + 1 UCCAGCUACCUGAAGUAGCUGAGCCGGGGCUCAACAAUGGCUCAAUUGUUCCUGUUCGCCACCGGUUCAAACGAGUUUCGGGCGGUAAGACGAGAUUCGUGGAUCUCCAGAACUCUGAAC (((((((((.....))))))((((((((((..((((..(((......)))..)))).))).)))))))..((((((((((..(.....).)))))))))))))....(((....)))... ( -46.20) >DroEre_CAF1 5701 119 + 1 UCCAGCUACCUGAAGUAGCUGAGCCGGAGCUCAACAAUGGCUCAAUUGUUCCUGUUCACCACCGGUUUAAACGAGUUUUGGGCGGCAAGACGAGAUUUCUAGAUCUCCAGAACUCUGAA- ..(((((((.....)))))))(((((((((..((((((......))))))...))).....)))))).....(((((((((.((......))(((((....))))))))))))))....- ( -37.40) >consensus UCCAGCUACCUGAAGUAGCUGAGCCGGAGCUCAACAAUGGCUCAAUUGUUCCUGUUCGCCACCGGUUCAAACGAGUUUCGGGCGGUAAGACGAGAUUCGUGGAUCUCCAGAACUCUGAAC ....(((((.....)))))(((((((((((..((((..(((......)))..)))).))).))))))))...((((((.((.((......))(((((....))))))).))))))..... (-32.74 = -33.43 + 0.69)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:20:37 2006