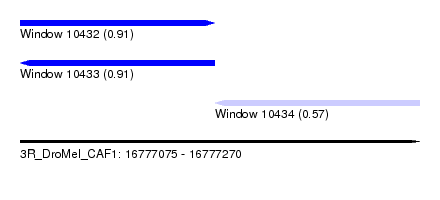
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,777,075 – 16,777,270 |
| Length | 195 |
| Max. P | 0.909495 |

| Location | 16,777,075 – 16,777,170 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 98.77 |
| Mean single sequence MFE | -26.75 |
| Consensus MFE | -26.20 |
| Energy contribution | -26.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.907682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
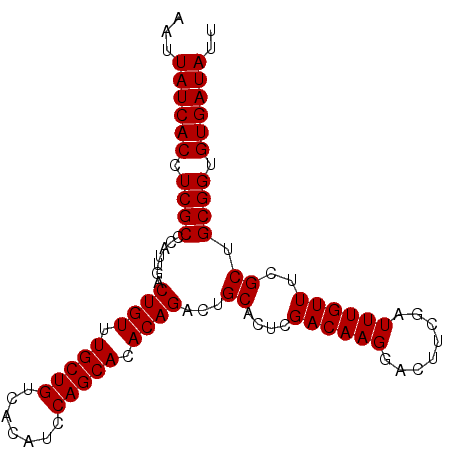
>3R_DroMel_CAF1 16777075 95 + 27905053 AAUUAUCACCUCGCCCAUUGACUGUUUGCUGUCACAUCCAGCACACAGAUUGCAUUCGACAAGGACUUCGAUUUGUUUCGCUGCGGUGUGAUAUU ...((((((.((((.......((((.(((((.......))))).))))...(((.((((........))))..)))......)))).)))))).. ( -26.40) >DroSec_CAF1 9475 95 + 1 AAUUAUCACCUCGCCCAUUGACUGUUUGCUGUCACAUCCAGCACACAGACUGCACUCGACAAGGACUUCGAUUUGUUUCGCUGCGGUGUGAUAUU ...((((((............((((.(((((.......))))).))))(((((((..((((((........))))))..).)))))))))))).. ( -27.20) >DroSim_CAF1 11122 95 + 1 AAUUAUCACCUCGCCCAUUGACUGUUUGCUGUCACAUCCAGCACACAGACUGCACUCGACAAGGACUUCGAUUUGUUUCGCUGCGGUGUGAUAUU ...((((((............((((.(((((.......))))).))))(((((((..((((((........))))))..).)))))))))))).. ( -27.20) >DroEre_CAF1 5296 95 + 1 AAUUAUCACCUCGCCCAUUGACUGUUUGCUGUCACAUCCAGCACACAGAUUGCACUCGACAAGGACUUCGAUUUGUUUCGCUGCGGUGUGAUAUU ...((((((.((((.......((((.(((((.......))))).))))...((....((((((........))))))..)).)))).)))))).. ( -26.20) >consensus AAUUAUCACCUCGCCCAUUGACUGUUUGCUGUCACAUCCAGCACACAGACUGCACUCGACAAGGACUUCGAUUUGUUUCGCUGCGGUGUGAUAUU ...((((((.((((.......((((.(((((.......))))).))))...((....((((((........))))))..)).)))).)))))).. (-26.20 = -26.20 + -0.00)

| Location | 16,777,075 – 16,777,170 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 98.77 |
| Mean single sequence MFE | -30.07 |
| Consensus MFE | -28.75 |
| Energy contribution | -28.75 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
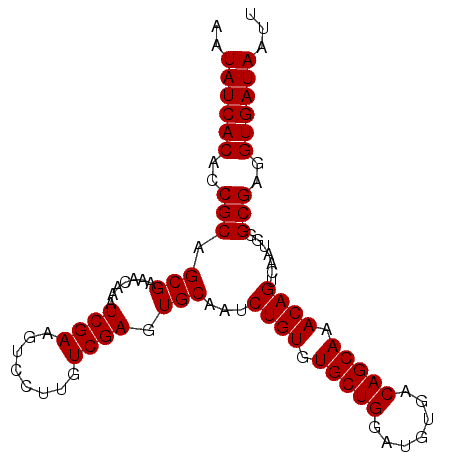
>3R_DroMel_CAF1 16777075 95 - 27905053 AAUAUCACACCGCAGCGAAACAAAUCGAAGUCCUUGUCGAAUGCAAUCUGUGUGCUGGAUGUGACAGCAAACAGUCAAUGGGCGAGGUGAUAAUU ..((((((...((..(((......)))..)).((((((.(.((....((((.(((((.......))))).)))).)).).))))))))))))... ( -28.60) >DroSec_CAF1 9475 95 - 1 AAUAUCACACCGCAGCGAAACAAAUCGAAGUCCUUGUCGAGUGCAGUCUGUGUGCUGGAUGUGACAGCAAACAGUCAAUGGGCGAGGUGAUAAUU ..((((((..(((.(((.......((((........)))).))).(.((((.(((((.......))))).)))).).....)))..))))))... ( -31.20) >DroSim_CAF1 11122 95 - 1 AAUAUCACACCGCAGCGAAACAAAUCGAAGUCCUUGUCGAGUGCAGUCUGUGUGCUGGAUGUGACAGCAAACAGUCAAUGGGCGAGGUGAUAAUU ..((((((..(((.(((.......((((........)))).))).(.((((.(((((.......))))).)))).).....)))..))))))... ( -31.20) >DroEre_CAF1 5296 95 - 1 AAUAUCACACCGCAGCGAAACAAAUCGAAGUCCUUGUCGAGUGCAAUCUGUGUGCUGGAUGUGACAGCAAACAGUCAAUGGGCGAGGUGAUAAUU ..((((((..(((.(((.......((((........)))).)))...((((.(((((.......))))).)))).......)))..))))))... ( -29.30) >consensus AAUAUCACACCGCAGCGAAACAAAUCGAAGUCCUUGUCGAGUGCAAUCUGUGUGCUGGAUGUGACAGCAAACAGUCAAUGGGCGAGGUGAUAAUU ..((((((..(((.(((.......((((........)))).)))...((((.(((((.......))))).)))).......)))..))))))... (-28.75 = -28.75 + 0.00)

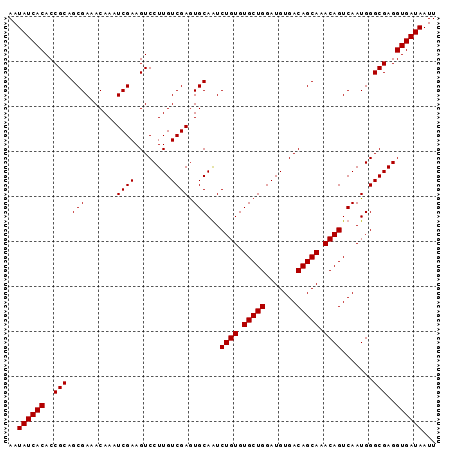
| Location | 16,777,170 – 16,777,270 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 93.14 |
| Mean single sequence MFE | -29.27 |
| Consensus MFE | -24.58 |
| Energy contribution | -24.20 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.568652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16777170 100 - 27905053 ACCGAUACCUGGGAAACUGGAGGCCCAGACCCCUAAUAGCGGGUUAUGCAAUCGUUAGUAAUUAAGACCCUACCCAUUGACGCAAACUGGGC----AUGUGCCA ...(.((((..(....)..)..(((((((((((.....).))))........(((((((................)))))))....))))))----..))).). ( -25.69) >DroSec_CAF1 9570 100 - 1 ACGGAUACCUGGGAAACUGGAGGCCCAGACCCCUAACACCGGGUUAUGCAAUCGUUAGUAAUUAAAACCCUAUCCAUUGACGCAAACUGGGC----AUGUGCCA ..((.((((..(....)..)..((((((((((........))))........(((((((...((......))...)))))))....))))))----..))))). ( -28.90) >DroSim_CAF1 11217 100 - 1 ACGGAUACCUGGGAAACUGGAGGCCCAGACCCCUAACAGCGGGUUAUGCAAUCGUUAGUAAUUAAAACCCUAUCCAUUGACGCAAACUGGGC----AUGUGCCA ..((.((((..(....)..)..(((((((((((.....).))))........(((((((...((......))...)))))))....))))))----..))))). ( -29.20) >DroEre_CAF1 5391 104 - 1 ACAGAUACCUGGGAAACUGGAGGCCCAGACCCCUAACAGUGGGUUAUGCAAUUGUCAGUAAUUAAAACCCUACCUAUUGACGCAAACUGGGCAGCCAUGUGCCA .(((....)))((....(((..(((((((((((.....).))))..(((....(((((((..((......))..))))))))))..))))))..)))....)). ( -33.30) >consensus ACGGAUACCUGGGAAACUGGAGGCCCAGACCCCUAACAGCGGGUUAUGCAAUCGUUAGUAAUUAAAACCCUACCCAUUGACGCAAACUGGGC____AUGUGCCA ...(.((((..(....)..)..((((((((((........))))........(((((((...((......))...)))))))....))))))......))).). (-24.58 = -24.20 + -0.38)

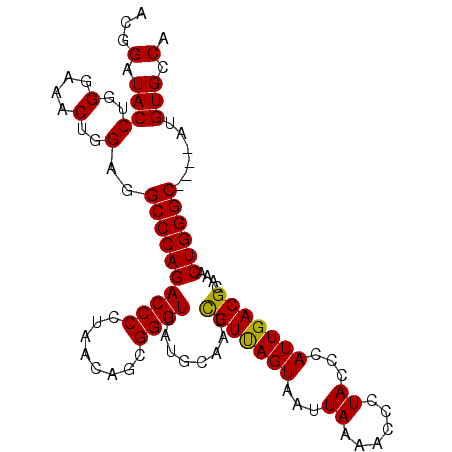
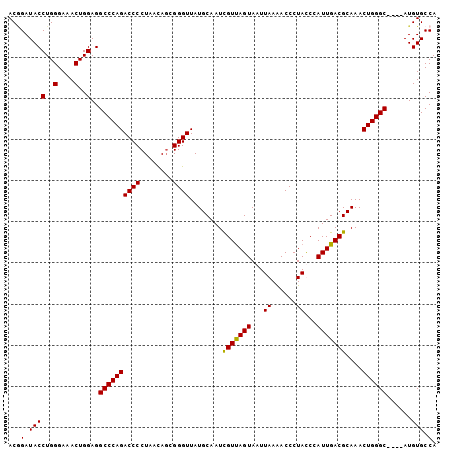
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:20:35 2006