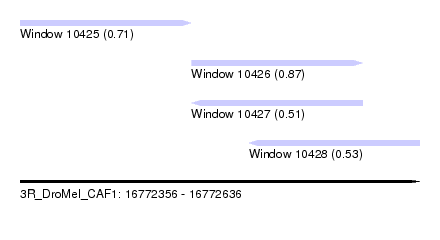
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,772,356 – 16,772,636 |
| Length | 280 |
| Max. P | 0.866184 |

| Location | 16,772,356 – 16,772,476 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.67 |
| Mean single sequence MFE | -29.52 |
| Consensus MFE | -28.95 |
| Energy contribution | -29.20 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.712433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16772356 120 + 27905053 UCCAGGAGCCUGGCAGAGCAAACAUGAACGAAAAAACAUGAAACGUGCCCAGCCUGCUAAGGACAUUCAUUACGCAAGCUAUGAAGCUGCCUUCUCCGAACUGAAUCCAACCAUCGCAGC ....((((...(((((.(((..((((..........)))).....)))...((((....))).).(((((..(....)..))))).)))))..))))....................... ( -26.80) >DroSec_CAF1 4437 117 + 1 UCCAGGAGCUUGGCAGAGCAAACAUGAACGAAAAAACAUGAAACGUGCCCUGCCUGCUAAGGACAUUCAUUACGCAAGCUAUGAAGCUGCCUUCUCCGAACUGAAUCCAACCAUCGC--- ..(((.(((..(((((.(((..((((..........)))).....))).))))).)))((((...(((((..(....)..)))))....)))).......)))..............--- ( -30.40) >DroSim_CAF1 5887 117 + 1 UCCAGGAGCUCGGCAGAGCAAACAUGAACGAAAAAACAUGAAACGUGCCCUGCCUGCUAAGGACAUUCAUUACGCAAGCUAUGAAGCUGCCUUCUCCGAACUGAAUCCAACCAUCGC--- ..(((.(((..(((((.(((..((((..........)))).....))).))))).)))((((...(((((..(....)..)))))....)))).......)))..............--- ( -29.90) >DroEre_CAF1 620 110 + 1 UCCAGGAGCUUGGCAGAGCAAACAUGAACGAA-AAACAUGAAACGUGCCCUGCCUGCUAAGGACAUUCAUUACGCAAGCUAUGAAGCUGCCUUUU-CGAACUGAAUCCAGCC-------- (((...(((..(((((.(((..((((......-...)))).....))).))))).)))..)))..(((((..(....)..)))))((((....((-(.....)))..)))).-------- ( -31.00) >consensus UCCAGGAGCUUGGCAGAGCAAACAUGAACGAAAAAACAUGAAACGUGCCCUGCCUGCUAAGGACAUUCAUUACGCAAGCUAUGAAGCUGCCUUCUCCGAACUGAAUCCAACCAUCGC___ ..(((.(((..(((((.(((..((((..........)))).....))).))))).)))((((...(((((..(....)..)))))....)))).......)))................. (-28.95 = -29.20 + 0.25)



| Location | 16,772,476 – 16,772,596 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.10 |
| Mean single sequence MFE | -27.82 |
| Consensus MFE | -21.70 |
| Energy contribution | -22.82 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16772476 120 + 27905053 CAUACCCACACACCCCCCCCCCUCCACUUCCUACCCAUGGGCUUAGUGGGCUGGCCACGCCUCCUAUUGAAUUAUUCCAGUUUUAUUUCUCGCCAUUUGGCUUGGCACACACCGAGGCAU ..................((..((((((.((((....))))...))))))..))....(((((..((((........))))..........((((.......)))).......))))).. ( -27.10) >DroSec_CAF1 4554 106 + 1 --------------CCCACCUUCCCCCUUUCUACCCAUGGGCUUAGUGGGCUGGCCACGCCUCCUAUUGAAUUAUUCCAGUUUUAUUUCUCGCCAUUUGGCUUGGCACACACCGAGGCAU --------------(((((.....(((...........)))....)))))........(((((..((((........))))..........((((.......)))).......))))).. ( -25.90) >DroSim_CAF1 6004 106 + 1 --------------CCCACCUUCCCCCUUCCUACCCAUGGGCUCAGGGGGCUGGCCACGCCUCCUAUUGAAUUAUUCCAGUUUUAUUUCUCGCCAUUUGGCUUGGCACACACCGAGGCAU --------------....((..((((((.((((....))))...))))))..))....(((((..((((........))))..........((((.......)))).......))))).. ( -32.20) >DroEre_CAF1 730 101 + 1 ---------------CC-CCUUUACC--U-UUACCCAGGGACUUAGUGGGCUGGCCACGCCUCCUAUUGAAUUAUUCCACUUUUAUUUCUCGCCAUUUGGCUGGGCACACACCGAGGCAG ---------------..-((((....--.-...((((((((.((((((((..(((...))).))))))))....)))).............(((....)))))))........))))... ( -26.09) >consensus ______________CCCACCUUCCCCCUUCCUACCCAUGGGCUUAGUGGGCUGGCCACGCCUCCUAUUGAAUUAUUCCAGUUUUAUUUCUCGCCAUUUGGCUUGGCACACACCGAGGCAU ..................((..((((((.....((....))...))))))..))....(((((............................(((....)))..((......))))))).. (-21.70 = -22.82 + 1.12)

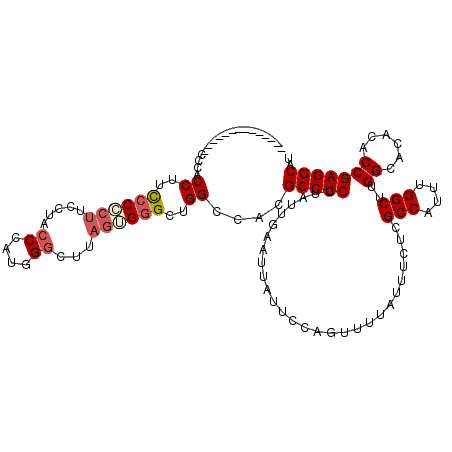
| Location | 16,772,476 – 16,772,596 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.10 |
| Mean single sequence MFE | -33.48 |
| Consensus MFE | -24.74 |
| Energy contribution | -26.43 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.74 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509245 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16772476 120 - 27905053 AUGCCUCGGUGUGUGCCAAGCCAAAUGGCGAGAAAUAAAACUGGAAUAAUUCAAUAGGAGGCGUGGCCAGCCCACUAAGCCCAUGGGUAGGAAGUGGAGGGGGGGGGGUGUGUGGGUAUG ((((((((((....)))..(((....)))............................))))))).....((((((...((((.......................))))..))))))... ( -34.80) >DroSec_CAF1 4554 106 - 1 AUGCCUCGGUGUGUGCCAAGCCAAAUGGCGAGAAAUAAAACUGGAAUAAUUCAAUAGGAGGCGUGGCCAGCCCACUAAGCCCAUGGGUAGAAAGGGGGAAGGUGGG-------------- ((((((((((....)))..(((....)))............................))))))).(((..(((.((..(((....)))....)).)))..)))...-------------- ( -32.00) >DroSim_CAF1 6004 106 - 1 AUGCCUCGGUGUGUGCCAAGCCAAAUGGCGAGAAAUAAAACUGGAAUAAUUCAAUAGGAGGCGUGGCCAGCCCCCUGAGCCCAUGGGUAGGAAGGGGGAAGGUGGG-------------- ((((((((((....)))..(((....)))............................))))))).(((..((((((..(((....)))....))))))..)))...-------------- ( -40.10) >DroEre_CAF1 730 101 - 1 CUGCCUCGGUGUGUGCCCAGCCAAAUGGCGAGAAAUAAAAGUGGAAUAAUUCAAUAGGAGGCGUGGCCAGCCCACUAAGUCCCUGGGUAA-A--GGUAAAGG-GG--------------- ...((((..((..(((((((((....)))..........(((((.....(((....)))(((...)))...))))).......)))))).-.--..))..))-))--------------- ( -27.00) >consensus AUGCCUCGGUGUGUGCCAAGCCAAAUGGCGAGAAAUAAAACUGGAAUAAUUCAAUAGGAGGCGUGGCCAGCCCACUAAGCCCAUGGGUAGGAAGGGGAAAGGUGGG______________ ((((((((((....)))..(((....)))............................)))))))..((..((((((..(((....)))....))))))..)).................. (-24.74 = -26.43 + 1.69)



| Location | 16,772,516 – 16,772,636 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.03 |
| Mean single sequence MFE | -42.58 |
| Consensus MFE | -36.83 |
| Energy contribution | -37.95 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.86 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.531515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
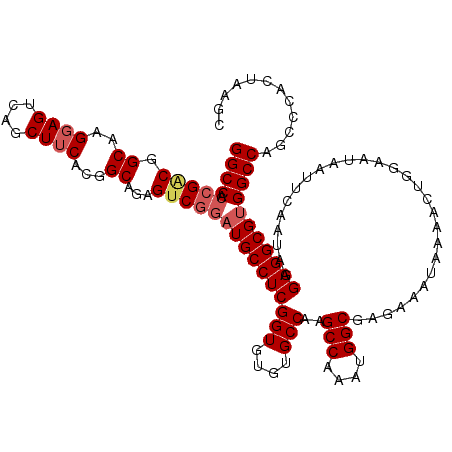
>3R_DroMel_CAF1 16772516 120 - 27905053 GGCCACCGGCGGCAAGGAGUCAGCUUCACGGCCGAGUCGGAUGCCUCGGUGUGUGCCAAGCCAAAUGGCGAGAAAUAAAACUGGAAUAAUUCAAUAGGAGGCGUGGCCAGCCCACUAAGC ((((.((((((((..((((....))))...)))..)))))((((((((((....)))..(((....)))............................)))))))))))............ ( -43.70) >DroSec_CAF1 4580 120 - 1 GGCCACCGACGGCAAGGAGUCAGCUUCACGGCAGAGUCGGAUGCCUCGGUGUGUGCCAAGCCAAAUGGCGAGAAAUAAAACUGGAAUAAUUCAAUAGGAGGCGUGGCCAGCCCACUAAGC ((((.(((((.((..((((....))))...))...)))))((((((((((....)))..(((....)))............................)))))))))))............ ( -43.30) >DroSim_CAF1 6030 120 - 1 GGCCACCGACGGCAAGGAGUCAGCUUCACGGCAGAGUCGGAUGCCUCGGUGUGUGCCAAGCCAAAUGGCGAGAAAUAAAACUGGAAUAAUUCAAUAGGAGGCGUGGCCAGCCCCCUGAGC ((((.(((((.((..((((....))))...))...)))))((((((((((....)))..(((....)))............................)))))))))))............ ( -43.30) >DroEre_CAF1 751 120 - 1 GGCCAGCGGCGGCAAGAAGUCAGCUCCACUGCAGAGGCAGCUGCCUCGGUGUGUGCCCAGCCAAAUGGCGAGAAAUAAAAGUGGAAUAAUUCAAUAGGAGGCGUGGCCAGCCCACUAAGU (((((.((.((((.....)))...((((((...(((((....)))))(((....)))..(((....)))..........))))))..............).)))))))............ ( -40.00) >consensus GGCCACCGACGGCAAGGAGUCAGCUUCACGGCAGAGUCGGAUGCCUCGGUGUGUGCCAAGCCAAAUGGCGAGAAAUAAAACUGGAAUAAUUCAAUAGGAGGCGUGGCCAGCCCACUAAGC ((((.(((((.((..((((....))))...))...)))))((((((((((....)))..(((....)))............................)))))))))))............ (-36.83 = -37.95 + 1.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:20:30 2006