
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,770,142 – 16,770,542 |
| Length | 400 |
| Max. P | 0.954673 |

| Location | 16,770,142 – 16,770,262 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.66 |
| Mean single sequence MFE | -30.03 |
| Consensus MFE | -23.92 |
| Energy contribution | -24.03 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.668661 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16770142 120 - 27905053 AAAACGCCUUUGGUGCAGCGUGGAAAACGGGAAACUUUUCCCUACCCGCCUAGAAAAACACUAAACAAAAGUUGAGUCGAGCAAAACAACAGAGGCAACAAACAUCGCGAAACAACAUUA .....(((((((.(((((.((((.....(((((....)))))...))))))........(((.(((....))).)))...)))......)))))))........................ ( -26.50) >DroSec_CAF1 2191 119 - 1 AAAACGCCUUUGGUGCGGCGUGGAAAGCGAGAAAGUUUUC-CUGCCCGCCUAGAAAAACACUAAACAAAAGUUGAGUCGAGCAAAACAACAGAGGCAACAAACAUCGCGAAACAACAUUA .....((((((((((.((((.(((((((......))))))-))))))))).........(((.(((....))).))).............))))))........................ ( -31.80) >DroSim_CAF1 3651 119 - 1 AAAACGCCUUUGGUGCGGCGUGGAAAGCGAGAAAGUUUUC-CUGCCCGCCUAGAAAAACACUAAACAAAAGUUGAGUCGAGCAAAACAACAGAGGCAACAAACAUCGCGAAACAACAUUA .....((((((((((.((((.(((((((......))))))-))))))))).........(((.(((....))).))).............))))))........................ ( -31.80) >consensus AAAACGCCUUUGGUGCGGCGUGGAAAGCGAGAAAGUUUUC_CUGCCCGCCUAGAAAAACACUAAACAAAAGUUGAGUCGAGCAAAACAACAGAGGCAACAAACAUCGCGAAACAACAUUA .....(((((((.(((((((.((.....((((....))))....)))))).........(((.(((....))).)))...)))......)))))))........................ (-23.92 = -24.03 + 0.11)

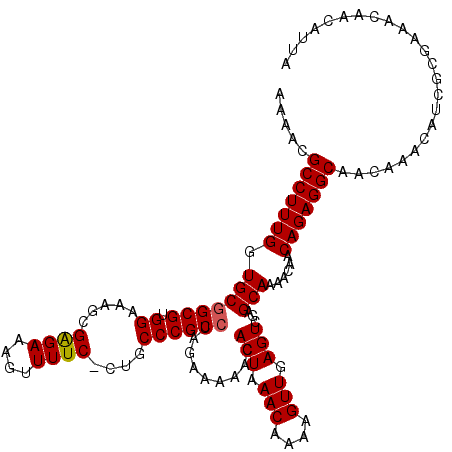

| Location | 16,770,182 – 16,770,302 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.87 |
| Mean single sequence MFE | -33.53 |
| Consensus MFE | -30.33 |
| Energy contribution | -29.45 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.743232 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16770182 120 + 27905053 UCGACUCAACUUUUGUUUAGUGUUUUUCUAGGCGGGUAGGGAAAAGUUUCCCGUUUUCCACGCUGCACCAAAGGCGUUUUCCCUUUUUUCUCGCACGAAACUGUUGAGUGGCAAGUACAC .(.(((((((....((((.((((.......((((((..(((((....))))).....)).)))).....(((((.(....))))))......)))).)))).))))))).)......... ( -32.90) >DroSec_CAF1 2231 119 + 1 UCGACUCAACUUUUGUUUAGUGUUUUUCUAGGCGGGCAG-GAAAACUUUCUCGCUUUCCACGCCGCACCAAAGGCGUUUUUCCCUUUUUCUCACACAAAACUGUUGAGUGGCAAGUACAC .(.(((((((....((((.((((.......((..(((..-(((....)))..)))..))(((((........)))))...............)))).)))).))))))).)......... ( -33.70) >DroSim_CAF1 3691 119 + 1 UCGACUCAACUUUUGUUUAGUGUUUUUCUAGGCGGGCAG-GAAAACUUUCUCGCUUUCCACGCCGCACCAAAGGCGUUUUUCCCUUUUCCUCGCACAAAACUGUUGAGUGGCAAGUACAC .(.(((((((....((((.((((.......((..(((..-(((....)))..)))..))(((((........)))))...............)))).)))).))))))).)......... ( -34.00) >consensus UCGACUCAACUUUUGUUUAGUGUUUUUCUAGGCGGGCAG_GAAAACUUUCUCGCUUUCCACGCCGCACCAAAGGCGUUUUUCCCUUUUUCUCGCACAAAACUGUUGAGUGGCAAGUACAC .(.(((((((....((((.((((......(((((((.............)))))))...(((((........)))))...............)))).)))).))))))).)......... (-30.33 = -29.45 + -0.88)



| Location | 16,770,262 – 16,770,382 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.11 |
| Mean single sequence MFE | -32.70 |
| Consensus MFE | -27.62 |
| Energy contribution | -27.73 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.794228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16770262 120 - 27905053 UGUGUGUUUGUGCUGGUGUUGAUGACUGCACAAAGCACACAAACUCGAAAAAAAGAACACACACACACGGCAACACUUCUGUGUACUUGCCACUCAACAGUUUCGUGCGAGAAAAAAGGG ((((((((((((((.((((........))))..))))).(......).......))))))))).....((((((((....))))...)))).(((.((......))..)))......... ( -32.20) >DroSec_CAF1 2310 112 - 1 ------UGUGUGCGGGUGUUGAUGACUGCACAAAGCACACAAACUCGAAAAAAAGAA--CACACACACGGCAACACUUCUGUGUACUUGCCACUCAACAGUUUUGUGUGAGAAAAAGGGA ------(((((((..((((........))))...)))))))................--..((((((.((((((((....))))...))))(((....)))..))))))........... ( -32.40) >DroSim_CAF1 3770 112 - 1 UA----UGUGUGCGGGUGUUGAUGACUGCACAAAGCACACAAACUCGAAAAAA--AA--CACACACACGGCAACACUUCUGUGUACUUGCCACUCAACAGUUUUGUGCGAGGAAAAGGGA ..----(((((((..((((........))))...)))))))..((((....((--((--(........((((((((....))))...))))........)))))...))))......... ( -33.49) >consensus U_____UGUGUGCGGGUGUUGAUGACUGCACAAAGCACACAAACUCGAAAAAAAGAA__CACACACACGGCAACACUUCUGUGUACUUGCCACUCAACAGUUUUGUGCGAGAAAAAGGGA ......(((((((..((((........))))...)))))))..((((.....................((((((((....))))...)))).....((......)).))))......... (-27.62 = -27.73 + 0.11)



| Location | 16,770,302 – 16,770,422 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.37 |
| Mean single sequence MFE | -37.35 |
| Consensus MFE | -31.83 |
| Energy contribution | -33.67 |
| Covariance contribution | 1.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.86 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.954673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16770302 120 - 27905053 UAAUGUCCUUUCUAUUUACUAUCCACUGGCUGUGUGUGCGUGUGUGUUUGUGCUGGUGUUGAUGACUGCACAAAGCACACAAACUCGAAAAAAAGAACACACACACACGGCAACACUUCU ............................((((((((((.((((((((((((((.(((.......))))))).))))))......((........)))))).))))))))))......... ( -36.40) >DroSec_CAF1 2350 112 - 1 UAAUGUCCUUUCUAUUUACUAUCCACUGGCUGUGUGUGUG------UGUGUGCGGGUGUUGAUGACUGCACAAAGCACACAAACUCGAAAAAAAGAA--CACACACACGGCAACACUUCU ............................((((((((((((------(((((((..((((........))))...))))))....((........)))--))))))))))))......... ( -38.90) >DroSim_CAF1 3810 112 - 1 UAAUGUCCUUUCUAUUUACUAUCCACUGGCUGUGUGUGUGUA----UGUGUGCGGGUGUUGAUGACUGCACAAAGCACACAAACUCGAAAAAA--AA--CACACACACGGCAACACUUCU ............................(((((((((((((.----(((((((..((((........))))...)))))))............--.)--))))))))))))......... ( -36.74) >consensus UAAUGUCCUUUCUAUUUACUAUCCACUGGCUGUGUGUGUGU_____UGUGUGCGGGUGUUGAUGACUGCACAAAGCACACAAACUCGAAAAAAAGAA__CACACACACGGCAACACUUCU ............................((((((((((((((....(((((((..((((........))))...)))))))...((........)).))))))))))))))......... (-31.83 = -33.67 + 1.83)



| Location | 16,770,422 – 16,770,542 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -43.17 |
| Consensus MFE | -40.97 |
| Energy contribution | -41.63 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.889037 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16770422 120 + 27905053 AGUGGCGUUAAACAGACACUCGUGCAACAGCCGCAACAUGUGAAGUGGACCAGCGGGGAAGUGGCAAGCCAGGACAUCGGGUGGUUGGGUGUGAAAAAAAUGCAUGAGGCUGCGCCAUUG (((((((.....(((...(((((((((((.((....(((.....)))(((((.(.((....(((....))).....)).).))))))).)))........)))))))).)))))))))). ( -40.90) >DroSec_CAF1 2462 120 + 1 AGUGGCGUUAAACAGACACUCGUGCAACAGCCGCAACAUGUGAAGUGGACCAGCGGGGAAGUGGCACGCCAGGACAUCGGGUGGAUUGGUGGGGAAAAAAGGCAUGAGGCUGCGCCAUUG (((((((.....(((...(((((((....(((((..(((.....)))..((....))...))))).((((((..(((...)))..))))))..........))))))).)))))))))). ( -44.30) >DroSim_CAF1 3922 120 + 1 AGUGGCGUUAAACAGACACUCGUGCAACAGCCGCAACAUGUGAAGUGGACCAGCGGGGAAGUGGCACGCCAGGACAUCGGGUGGAUUGGUGGGGAAAAAAGGCAUGAGGCUGCGCCAUUG (((((((.....(((...(((((((....(((((..(((.....)))..((....))...))))).((((((..(((...)))..))))))..........))))))).)))))))))). ( -44.30) >consensus AGUGGCGUUAAACAGACACUCGUGCAACAGCCGCAACAUGUGAAGUGGACCAGCGGGGAAGUGGCACGCCAGGACAUCGGGUGGAUUGGUGGGGAAAAAAGGCAUGAGGCUGCGCCAUUG (((((((.....(((...(((((((....(((((..(((.....)))..((....))...))))).((((((..(((...)))..))))))..........))))))).)))))))))). (-40.97 = -41.63 + 0.67)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:20:24 2006