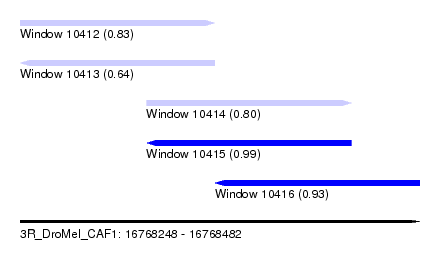
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,768,248 – 16,768,482 |
| Length | 234 |
| Max. P | 0.986995 |

| Location | 16,768,248 – 16,768,362 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.84 |
| Mean single sequence MFE | -41.37 |
| Consensus MFE | -37.72 |
| Energy contribution | -37.50 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829040 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
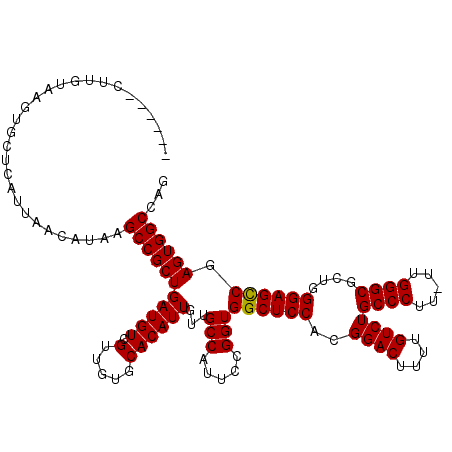
>3R_DroMel_CAF1 16768248 114 + 27905053 ------CUUGUAAGUGCUCAUUAACAUAAGCCGCUGAUGUCGUUGCGCACAUUGUUGCCAUUCCGGUGGCUCCACGGACUUUUGUCUGCCCUUUUUGGGCGCUGGGAGUCGAGUGGCCCG ------.......((((.((...((((.((...)).))))...)).))))......(((((((....((((((..((((....))))((((.....))))....)))))))))))))... ( -40.10) >DroSec_CAF1 1 119 + 1 UUACAGCUUGUAAGUGCUCAUUAACAUAAGCCGCUGAUGUCGUUGUGCACAUUGUUGCCAUUCCGGUGGCUCCACGGACUUUUGUCUGCCCUU-UUGGGCGCUGGGAGCCGAGUGGCCAG ((((.....))))((((.((...((((.((...)).))))...)).))))......(((((((....((((((..((((....))))((((..-..))))....)))))))))))))... ( -44.00) >DroSim_CAF1 1 99 + 1 --------------------UUAACAUAAGCCGCUGAUGUCGUUGUGCACAUUGUUGCCAUUCCGGUGGCUCCGCGGACUUUUGUCUGCCCUU-UUGGGCGCUGGGAGCCGAGUGGCCAG --------------------.........((((((....(((..(((((......)).)))..)))(((((((((((((....))))((((..-..))))))..)))))))))))))... ( -40.00) >consensus ______CUUGUAAGUGCUCAUUAACAUAAGCCGCUGAUGUCGUUGUGCACAUUGUUGCCAUUCCGGUGGCUCCACGGACUUUUGUCUGCCCUU_UUGGGCGCUGGGAGCCGAGUGGCCAG .............................(((((((((((.(.....))))))...(((.....)))((((((..((((....))))((((.....))))....)))))).))))))... (-37.72 = -37.50 + -0.22)

| Location | 16,768,248 – 16,768,362 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.84 |
| Mean single sequence MFE | -34.87 |
| Consensus MFE | -30.59 |
| Energy contribution | -30.70 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.642176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
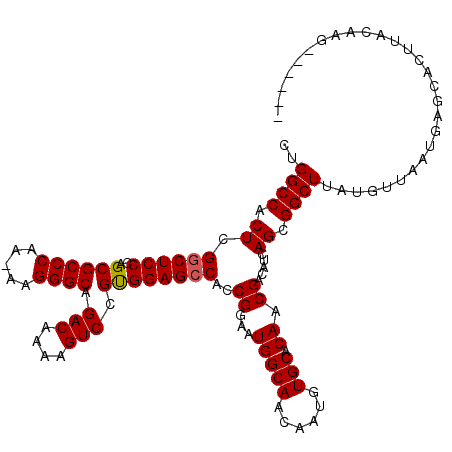
>3R_DroMel_CAF1 16768248 114 - 27905053 CGGGCCACUCGACUCCCAGCGCCCAAAAAGGGCAGACAAAAGUCCGUGGAGCCACCGGAAUGGCAACAAUGUGCGCAACGACAUCAGCGGCUUAUGUUAAUGAGCACUUACAAG------ (((.((((..((((.....(((((.....)))).).....)))).)))).....)))...((....)).(((((((..........)))((((((....))))))...))))..------ ( -32.20) >DroSec_CAF1 1 119 - 1 CUGGCCACUCGGCUCCCAGCGCCCAA-AAGGGCAGACAAAAGUCCGUGGAGCCACCGGAAUGGCAACAAUGUGCACAACGACAUCAGCGGCUUAUGUUAAUGAGCACUUACAAGCUGUAA ...((((.(((((((((...((((..-..)))).(((....))).).))))))....)).))))....((((........))))((((.((((((....))))))........))))... ( -38.60) >DroSim_CAF1 1 99 - 1 CUGGCCACUCGGCUCCCAGCGCCCAA-AAGGGCAGACAAAAGUCCGCGGAGCCACCGGAAUGGCAACAAUGUGCACAACGACAUCAGCGGCUUAUGUUAA-------------------- ..((((.((.((((((..((((((..-..)))).(((....))).))))))))..((...(((((......))).)).)).....)).))))........-------------------- ( -33.80) >consensus CUGGCCACUCGGCUCCCAGCGCCCAA_AAGGGCAGACAAAAGUCCGUGGAGCCACCGGAAUGGCAACAAUGUGCACAACGACAUCAGCGGCUUAUGUUAAUGAGCACUUACAAG______ ..((((.((.((((((..((((((.....)))).(((....))).))))))))..((...(((((......))).)).)).....)).))))............................ (-30.59 = -30.70 + 0.11)


| Location | 16,768,322 – 16,768,442 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.77 |
| Mean single sequence MFE | -42.93 |
| Consensus MFE | -38.59 |
| Energy contribution | -38.37 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.02 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.803001 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16768322 120 + 27905053 UUUGUCUGCCCUUUUUGGGCGCUGGGAGUCGAGUGGCCCGGAGUCCGGACUUGGGGCUCUCGACUCGCGCUGAUUGUCGUAUUUAAAUUAAUUUCAAUACAACGACAAAGAGUACGAAUC ((((((.((((.....))))((.(.((((((((.(((((.((((....)))).))))))))))))).))).....((.(((((.(((....))).))))).))))))))........... ( -49.70) >DroSec_CAF1 81 119 + 1 UUUGUCUGCCCUU-UUGGGCGCUGGGAGCCGAGUGGCCAGGAGUCCGGACUUGGGGCUCUCGACUCGCGCUGAUUGUCGUAUUUAAAUUAAUUUCAAUACAACAACAAAGAGUACGAAUC (((((((((((..-..))))((.(.(((.((((.((((..((((....))))..)))))))).))).)))((.((((.(((((.(((....))).))))).))))))...)).))))).. ( -39.10) >DroSim_CAF1 61 119 + 1 UUUGUCUGCCCUU-UUGGGCGCUGGGAGCCGAGUGGCCAGGAGUCCGGACUUGGGGCUCUCGACUCGCGCUGAUUGUCGUAUUUAAAUUAAUUUCAAUACAACGACAAAGAGUACGAAUC ((((((.((((..-..))))((.(.(((.((((.((((..((((....))))..)))))))).))).))).....((.(((((.(((....))).))))).))))))))........... ( -40.00) >consensus UUUGUCUGCCCUU_UUGGGCGCUGGGAGCCGAGUGGCCAGGAGUCCGGACUUGGGGCUCUCGACUCGCGCUGAUUGUCGUAUUUAAAUUAAUUUCAAUACAACGACAAAGAGUACGAAUC (((((((((((.....))))((.(.(((.((((.((((..((((....))))..)))))))).))).)))((.((((.(((((.(((....))).))))).))))))...)).))))).. (-38.59 = -38.37 + -0.22)

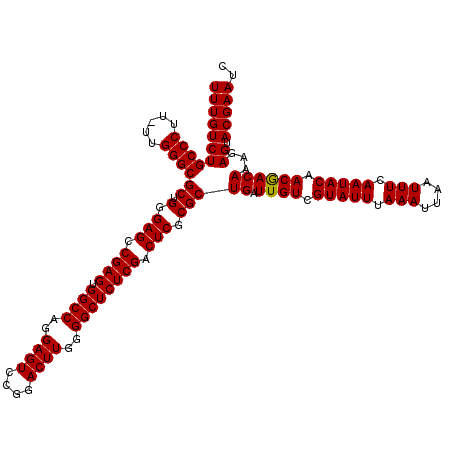
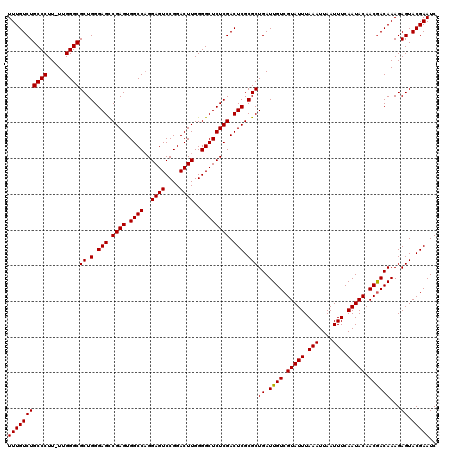
| Location | 16,768,322 – 16,768,442 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.77 |
| Mean single sequence MFE | -42.90 |
| Consensus MFE | -41.11 |
| Energy contribution | -40.67 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.65 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16768322 120 - 27905053 GAUUCGUACUCUUUGUCGUUGUAUUGAAAUUAAUUUAAAUACGACAAUCAGCGCGAGUCGAGAGCCCCAAGUCCGGACUCCGGGCCACUCGACUCCCAGCGCCCAAAAAGGGCAGACAAA ............(((((((((((((.(((....))).)))))))).....((..((((((((.((((..(((....)))..))))..))))))))...))((((.....)))).))))). ( -46.20) >DroSec_CAF1 81 119 - 1 GAUUCGUACUCUUUGUUGUUGUAUUGAAAUUAAUUUAAAUACGACAAUCAGCGCGAGUCGAGAGCCCCAAGUCCGGACUCCUGGCCACUCGGCUCCCAGCGCCCAA-AAGGGCAGACAAA ((((((..((....(((((((((((.(((....))).))))))))))).))..))))))(.(((((...(((((((....))))..))).))))).)...((((..-..))))....... ( -40.90) >DroSim_CAF1 61 119 - 1 GAUUCGUACUCUUUGUCGUUGUAUUGAAAUUAAUUUAAAUACGACAAUCAGCGCGAGUCGAGAGCCCCAAGUCCGGACUCCUGGCCACUCGGCUCCCAGCGCCCAA-AAGGGCAGACAAA ((((((..((..(((((((....(((((.....)))))..)))))))..))..))))))(.(((((...(((((((....))))..))).))))).)...((((..-..))))....... ( -41.60) >consensus GAUUCGUACUCUUUGUCGUUGUAUUGAAAUUAAUUUAAAUACGACAAUCAGCGCGAGUCGAGAGCCCCAAGUCCGGACUCCUGGCCACUCGGCUCCCAGCGCCCAA_AAGGGCAGACAAA ............(((((((((((((.(((....))).)))))))).....((..((((((((.(((...(((....)))...)))..))))))))...))((((.....)))).))))). (-41.11 = -40.67 + -0.44)

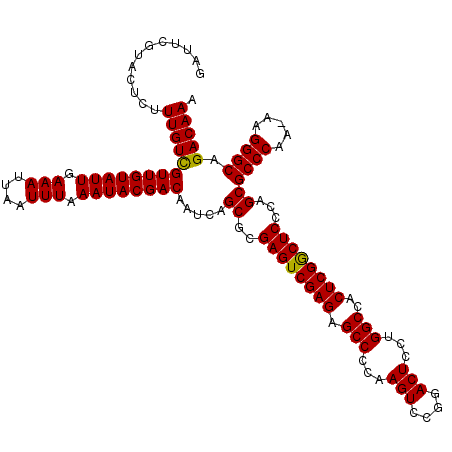
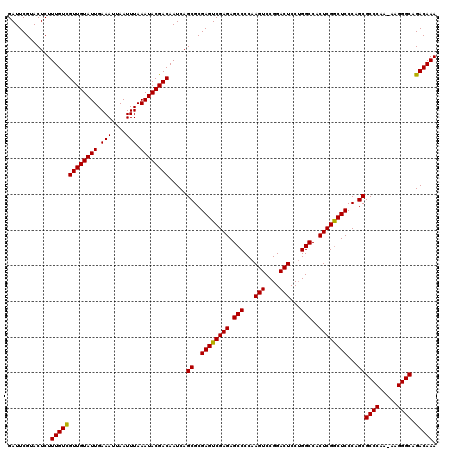
| Location | 16,768,362 – 16,768,482 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -32.33 |
| Consensus MFE | -31.85 |
| Energy contribution | -31.97 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16768362 120 - 27905053 ACUUUUAUGGGCCAGUGUCUUUUGAUUCUAUAAUUUGCAUGAUUCGUACUCUUUGUCGUUGUAUUGAAAUUAAUUUAAAUACGACAAUCAGCGCGAGUCGAGAGCCCCAAGUCCGGACUC ........(((((((.(((....))).))..........(((((((..((..(((((((....(((((.....)))))..)))))))..))..))))))).).))))............. ( -30.10) >DroSec_CAF1 120 120 - 1 ACUUUUAUGGGCCAGUGUCUUUUGAUUCUAUAAUUUGCUUGAUUCGUACUCUUUGUUGUUGUAUUGAAAUUAAUUUAAAUACGACAAUCAGCGCGAGUCGAGAGCCCCAAGUCCGGACUC ........((((.((.(((....))).))........(((((((((..((....(((((((((((.(((....))).))))))))))).))..))))))))).))))............. ( -33.10) >DroSim_CAF1 100 120 - 1 ACUUUUAUGGGCCAGUGUCUUUUGAUUCUAUAAUUUGCUUGAUUCGUACUCUUUGUCGUUGUAUUGAAAUUAAUUUAAAUACGACAAUCAGCGCGAGUCGAGAGCCCCAAGUCCGGACUC ........((((.((.(((....))).))........(((((((((..((..(((((((....(((((.....)))))..)))))))..))..))))))))).))))............. ( -33.80) >consensus ACUUUUAUGGGCCAGUGUCUUUUGAUUCUAUAAUUUGCUUGAUUCGUACUCUUUGUCGUUGUAUUGAAAUUAAUUUAAAUACGACAAUCAGCGCGAGUCGAGAGCCCCAAGUCCGGACUC ........((((.((.(((....))).))........(((((((((..((..(((((((....(((((.....)))))..)))))))..))..))))))))).))))............. (-31.85 = -31.97 + 0.11)

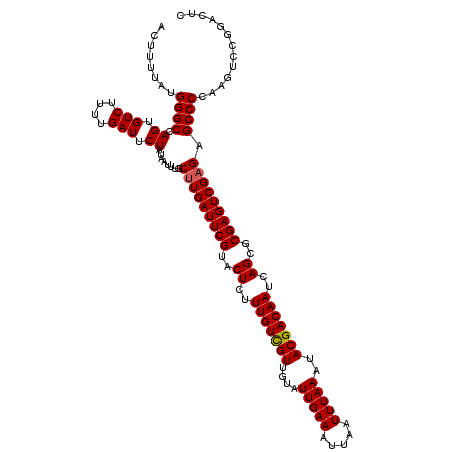
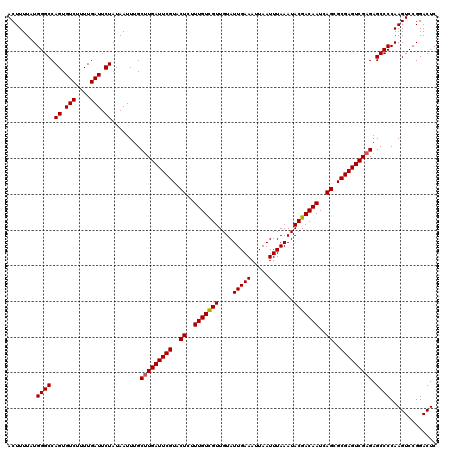
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:20:19 2006