
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,753,484 – 16,753,620 |
| Length | 136 |
| Max. P | 0.780065 |

| Location | 16,753,484 – 16,753,592 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 91.81 |
| Mean single sequence MFE | -27.33 |
| Consensus MFE | -25.35 |
| Energy contribution | -25.13 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.780065 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
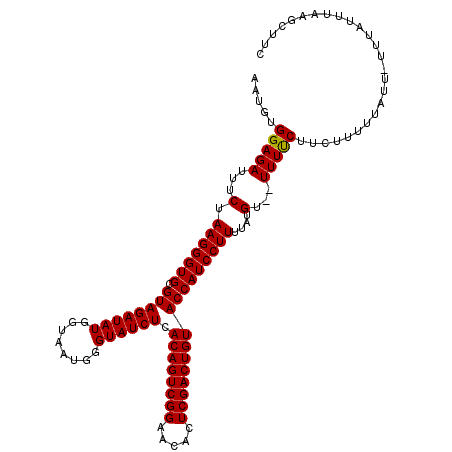
>3R_DroMel_CAF1 16753484 108 + 27905053 AAUGUGGAGAUUUCUAAGGGUGCGUAGAUAUGGUAAUGGGUAUCUCACAGUCGGGACACUCGACUGUACCAUCCUUUUAUGUGUUUUUCUUCUU-------UUUAUUCAAGCUUC ((((..((((.....(((((..((((((.((((((.((((...))))((((((((...))))))))))))))...))))))..)))))..))))-------..))))........ ( -28.30) >DroSec_CAF1 55938 112 + 1 AAUGCGGAGAUUUCUAAGGGUGCGUAGAUAUGGUAAUGGGUAUCUCACAGUCGGAACACUCGACUGUACCAUCCUUUUAUGU--UUUCCUUCUUUUUAUU-UUUAUUUAAGCUUC ...(((((((.....(((((((.((((((((........)))))).((((((((.....)))))))))))))))))......--)))))......(((..-......)))))... ( -28.00) >DroSim_CAF1 49353 113 + 1 AAUGUGGAGAUUUCUAAGGGUGCGUAGAUAUGGUAAUGGGUAUCUUACAGUCGGAACACUCGACUGUACCAUCCUUUUAUGU--UUUUCUUCUUUUUAUUUUUUAUUUAAGCUUC .....(((((...(.(((((((.((((((((........)))))).((((((((.....)))))))))))))))))....).--...)))))....................... ( -25.70) >consensus AAUGUGGAGAUUUCUAAGGGUGCGUAGAUAUGGUAAUGGGUAUCUCACAGUCGGAACACUCGACUGUACCAUCCUUUUAUGU__UUUUCUUCUUUUUAUU_UUUAUUUAAGCUUC .....(((((...(.(((((((.((((((((........)))))).((((((((.....)))))))))))))))))....)...))))).......................... (-25.35 = -25.13 + -0.22)


| Location | 16,753,524 – 16,753,620 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 91.50 |
| Mean single sequence MFE | -18.13 |
| Consensus MFE | -14.70 |
| Energy contribution | -14.48 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.703472 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
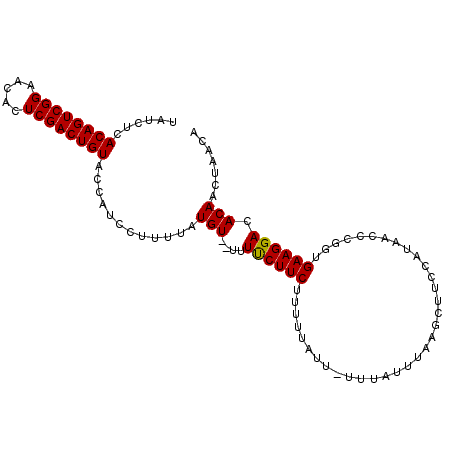
>3R_DroMel_CAF1 16753524 96 + 27905053 UAUCUCACAGUCGGGACACUCGACUGUACCAUCCUUUUAUGUGUUUUUCUUCUU-------UUUAUUCAAGCUUCCAUAACCCGGUGAAGGACACAACUAACA ......(((((((((...)))))))))............((((((((((.((..-------.((((..........))))...)).))))))))))....... ( -21.30) >DroSec_CAF1 55978 100 + 1 UAUCUCACAGUCGGAACACUCGACUGUACCAUCCUUUUAUGU--UUUCCUUCUUUUUAUU-UUUAUUUAAGCUUCCAUAACCCGGUGAAGGACACAACUAACA ......((((((((.....))))))))............(((--..((((((..((((..-......))))...((.......)).)))))).)))....... ( -17.30) >DroSim_CAF1 49393 101 + 1 UAUCUUACAGUCGGAACACUCGACUGUACCAUCCUUUUAUGU--UUUUCUUCUUUUUAUUUUUUAUUUAAGCUUCCAUAACCCGGUGAAGGACACAACUAACA .....(((((((((.....)))))))))...........(((--(((((..(((..((.....))...)))...((.......)).))))))))......... ( -15.80) >consensus UAUCUCACAGUCGGAACACUCGACUGUACCAUCCUUUUAUGU__UUUUCUUCUUUUUAUU_UUUAUUUAAGCUUCCAUAACCCGGUGAAGGACACAACUAACA ......((((((((.....))))))))............(((....((((((..................................)))))).)))....... (-14.70 = -14.48 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:20:03 2006