
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,715,728 – 16,715,921 |
| Length | 193 |
| Max. P | 0.997673 |

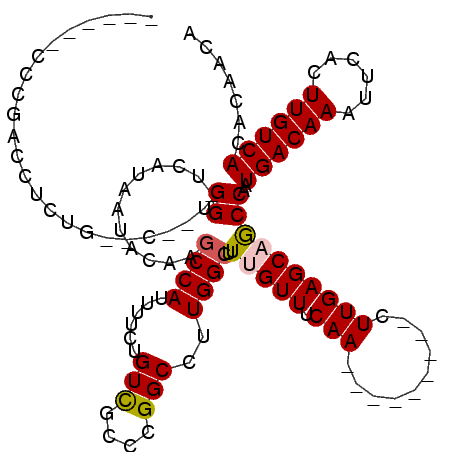
| Location | 16,715,728 – 16,715,827 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.50 |
| Mean single sequence MFE | -24.85 |
| Consensus MFE | -10.40 |
| Energy contribution | -10.60 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.42 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16715728 99 - 27905053 ------CCCGACCUCAG-----UGG-ACAAACUACAAGCCAUUUUCUGUCGACCGGCCUUGGCUUUGUUUCAA---------CUUGAGCAGCCAAUGACAAAUUCACUUGUCACACAACA ------..((((...((-----(((-.(.........))))))....))))...(((....((((.((....)---------)..)))).)))..((((((......))))))....... ( -20.40) >DroPse_CAF1 32533 105 - 1 ------CCCUACCACUGCCCGUUGGCUCGUAAUACAUGCCAUUUUCAGUCUCCCGGCCUUGGCUUUGUUUCAA---------CUUGAGCCACCAGUGACAAAUUCACUUGUCACACAACA ------..............(.(((((((....(((.((((......(((....)))..))))..))).....---------..))))))).).(((((((......)))))))...... ( -27.00) >DroYak_CAF1 43505 99 - 1 ------CCCGACCUCAU-----CGG-ACAAAAUACAAGCCAUUUUCUGUCGUCCGGCCUUGGCUUUGUUUCAA---------CUUGAGCAGCCAAUGACAAAUUCACUUGUCACACAACA ------.((((.....)-----)))-.......(((((........((((((..(((....((((.((....)---------)..)))).))).))))))......)))))......... ( -19.74) >DroAna_CAF1 28314 111 - 1 UACCAACCCCACCUCUGUUC--UGG-UCAUAC------CCAUUAGAUGUUGGGUGGCCUUGGCAUUGUUUCAAACCAGGCAGCUUGAGCAGCCAAUGACAAAUUCACUUGUCACACAACA ..((((..((((((....((--(((-(.....------..))))))....))))))..))))..((((.........(((.((....)).)))..((((((......)))))).)))).. ( -30.10) >DroPer_CAF1 31683 105 - 1 ------CCCUACCACUGCUCGUUGGCUCGUAAUACAUGCCAUUUUCAGUCUCCCGGCCUUGGCUUUGUUUCAA---------CUUGAGCCACCAGUGACAAAUUCACUUGUCACACAACA ------..............(.(((((((....(((.((((......(((....)))..))))..))).....---------..))))))).).(((((((......)))))))...... ( -27.00) >consensus ______CCCGACCUCUG__C__UGG_UCAUAAUACAAGCCAUUUUCUGUCGCCCGGCCUUGGCUUUGUUUCAA_________CUUGAGCAGCCAAUGACAAAUUCACUUGUCACACAACA .......................((............((((......(((....)))..)))).(((((.(((..........))))))))))..((((((......))))))....... (-10.40 = -10.60 + 0.20)


| Location | 16,715,827 – 16,715,921 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.69 |
| Mean single sequence MFE | -31.80 |
| Consensus MFE | -20.74 |
| Energy contribution | -21.14 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.995517 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16715827 94 + 27905053 ---AGAAAUUUGAAUUUAUGUUGGGGCC----------CGCA--------AA----AAGGGAAAUACUUUCAUUUGGAGC-UGCCACAAAGCAUUUCCGCUUUGUGGCACACACAUAAUG ---............((((((((.((((----------((..--------..----((((......))))....))).))-)((((((((((......)))))))))).)).)))))).. ( -29.70) >DroPse_CAF1 32638 109 + 1 ---GAAAAUUUGAAUCCAUGUUUUGGGCACUUU---UUUGCACUAGCUGGAA----AAGGAAAAUGCUUUCAUUUGGAGC-UGCCACAAAGCAUUUCCGCUUUGUGGCACACACAUAAUG ---...........((((......(((((((((---(((((....)).))))----))).....))))).....))))(.-(((((((((((......)))))))))))).......... ( -31.30) >DroYak_CAF1 43604 94 + 1 ---UGAAAUUUGAAUUUAUGUUGGGGGC----------CGCA--------AA----AAGGGAAAUACUUUCAUUUGGAGC-UGCCACAAAGCAUUUCCGCUUUGUGGCACACACAUAAUG ---............((((((((..(((----------..((--------((----((((......))))..))))..))-)((((((((((......)))))))))).)).)))))).. ( -28.40) >DroAna_CAF1 28425 112 + 1 UGGGAACAUUUGAAUUUAUGUUAGGACUAGUUUUCUUUCGCA--------UGGGUGGGGGAGAAUACUUUCAUAUGGAGCCUGCCACAAAGCAUUUCCGCUUUGUGGCACACACAUAAUG .(((..(((.((((.....((....))..(((((((..(((.--------...)))..)))))))...)))).)))...)))((((((((((......))))))))))............ ( -37.40) >DroPer_CAF1 31788 109 + 1 ---GAAAAUUUGAAUCCAUGUUUUGGGCACUUU---UUUGCACUAGCUGGAA----AAGGGAAAUGCUUUCAUUUGGAGC-UGCCACAAAGCAUUUCCGCUUUGUGGCACACACAUAAUG ---...........((((......(((((((((---(((.((.....)).))----)))))...))))).....))))(.-(((((((((((......)))))))))))).......... ( -32.20) >consensus ___GAAAAUUUGAAUUUAUGUUGGGGGCA_UUU___UUCGCA________AA____AAGGGAAAUACUUUCAUUUGGAGC_UGCCACAAAGCAUUUCCGCUUUGUGGCACACACAUAAUG ...............((((((......................................((((....))))..........(((((((((((......)))))))))))...)))))).. (-20.74 = -21.14 + 0.40)

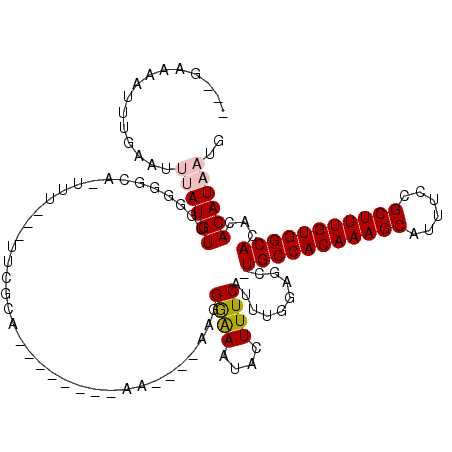

| Location | 16,715,827 – 16,715,921 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.69 |
| Mean single sequence MFE | -28.70 |
| Consensus MFE | -25.25 |
| Energy contribution | -24.53 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.91 |
| SVM RNA-class probability | 0.997673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16715827 94 - 27905053 CAUUAUGUGUGUGCCACAAAGCGGAAAUGCUUUGUGGCA-GCUCCAAAUGAAAGUAUUUCCCUU----UU--------UGCG----------GGCCCCAACAUAAAUUCAAAUUUCU--- ..((((((...((((((((((((....))))))))))))-((((((((.(((.....)))....----))--------)).)----------)))....))))))............--- ( -29.90) >DroPse_CAF1 32638 109 - 1 CAUUAUGUGUGUGCCACAAAGCGGAAAUGCUUUGUGGCA-GCUCCAAAUGAAAGCAUUUUCCUU----UUCCAGCUAGUGCAAA---AAAGUGCCCAAAACAUGGAUUCAAAUUUUC--- .....((.(.(((((((((((((....))))))))))))-.).)))..((((.(((((((..((----(....((....)))))---)))))))(((.....))).)))).......--- ( -27.60) >DroYak_CAF1 43604 94 - 1 CAUUAUGUGUGUGCCACAAAGCGGAAAUGCUUUGUGGCA-GCUCCAAAUGAAAGUAUUUCCCUU----UU--------UGCG----------GCCCCCAACAUAAAUUCAAAUUUCA--- ..((((((...((((((((((((....))))))))))))-(((.((((.(((.....)))....----))--------)).)----------)).....))))))............--- ( -27.80) >DroAna_CAF1 28425 112 - 1 CAUUAUGUGUGUGCCACAAAGCGGAAAUGCUUUGUGGCAGGCUCCAUAUGAAAGUAUUCUCCCCCACCCA--------UGCGAAAGAAAACUAGUCCUAACAUAAAUUCAAAUGUUCCCA ..((((((((.((((((((((((....)))))))))))).))..........(((.((((..(.((....--------)).)..)))).))).......))))))............... ( -30.60) >DroPer_CAF1 31788 109 - 1 CAUUAUGUGUGUGCCACAAAGCGGAAAUGCUUUGUGGCA-GCUCCAAAUGAAAGCAUUUCCCUU----UUCCAGCUAGUGCAAA---AAAGUGCCCAAAACAUGGAUUCAAAUUUUC--- .....((.(.(((((((((((((....))))))))))))-.).)))..((((.((((((...((----((...((....)))))---)))))))(((.....))).)))).......--- ( -27.60) >consensus CAUUAUGUGUGUGCCACAAAGCGGAAAUGCUUUGUGGCA_GCUCCAAAUGAAAGUAUUUCCCUU____UU________UGCGAA___AAA_UGCCCCAAACAUAAAUUCAAAUUUUC___ ..((((((((.((((((((((((....)))))))))))).))...........(((......................)))..................))))))............... (-25.25 = -24.53 + -0.72)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:19:50 2006