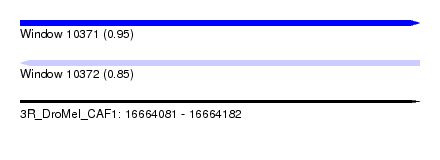
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,664,081 – 16,664,182 |
| Length | 101 |
| Max. P | 0.953016 |

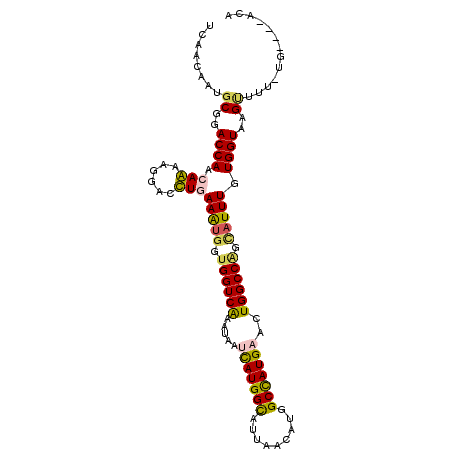
| Location | 16,664,081 – 16,664,182 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 79.55 |
| Mean single sequence MFE | -29.20 |
| Consensus MFE | -22.17 |
| Energy contribution | -21.82 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953016 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16664081 101 + 27905053 UCAACAAUGCAGACCAGCAGAAAGACCUAAAAUGGUGGUCGAACAAUCAUGGCAUUAACAUGGCCAUGAAUUGGCCAUCAUUUGUGGUGAGUUUUCUG----UUU ...............((((((((((((..(((((((((((((....(((((((.........))))))).)))))))))))))..)))...)))))))----)). ( -36.10) >DroVir_CAF1 7510 104 + 1 UUAACAAUGCGGACCAACAAAAGGACUUGAAAUGGUGGUCAAAUAAUCAUGGCAUAAAUAUGGCCAUGAAUUGGCCAGCAUUUGUGGUAAGUCUU-AGUCGGAAA .............((.((...(((((((((((((.(((((((....(((((((.........))))))).))))))).)))))....))))))))-.)).))... ( -32.40) >DroGri_CAF1 8053 98 + 1 UUAACAAUGCGGACCAACAAAAGGACUUGAAAUGGUGGUCAAAUAAUCAUGGCAUUAACAUGGCCAUGAACUGGCCGGCAUUUGUGGUAAGAG-------AGACA .........(..((((.(((......)))(((((.((((((.....(((((((.........)))))))..)))))).))))).))))..)..-------..... ( -26.20) >DroMoj_CAF1 6845 104 + 1 UCAACAAUGCGGACCAACAAAAGGACUUGAAAUGGUGGUCAAAUAACCAUGGUAUCAAUAUGGCCAUGAACUGGCCUGCAUUUGUGGUAAGUGUUUAGAUGG-UA .....(((((..((((.((((.....((((.((.(((((......))))).)).))))...(((((.....)))))....))))))))..))))).......-.. ( -27.80) >DroAna_CAF1 6320 100 + 1 UCAACAAUGCGGACCAGCAGAAAGACCUCAAAUUUUGGUCAAAUAAAUAUGGUAUUAACAUGGCUAUGAACUGGCCGGCAUUUGUGGUGAGUU-UCUG----ACU ...............((((((((.(((.(((((.(((((((.(((..((((.......))))..)))....))))))).))))).)))...))-))))----.)) ( -24.30) >DroPer_CAF1 8061 101 + 1 UCAACAAUGCGGACCAGCAGAAGGAUCUGAAGUGGUGGUCAAACAAUCAUGGUAUUAACAUGGCCAUGAACUGGCCAAUCUUUGUGGUAAGCUGUGUG----CCC ...(((..((..((((.((((....))))(((...((((((.....(((((((.........)))))))..))))))..)))..))))..))..))).----... ( -28.40) >consensus UCAACAAUGCGGACCAACAAAAGGACCUGAAAUGGUGGUCAAAUAAUCAUGGCAUUAACAUGGCCAUGAACUGGCCAGCAUUUGUGGUAAGUUUU_UG____ACA ........((..((((.(((......)))(((((.((((((.....(((((((.........)))))))..)))))).))))).))))..))............. (-22.17 = -21.82 + -0.36)

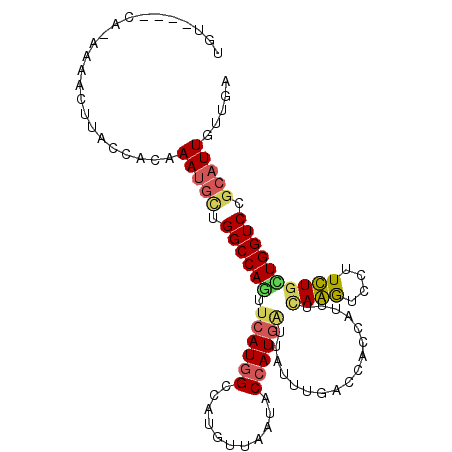
| Location | 16,664,081 – 16,664,182 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 79.55 |
| Mean single sequence MFE | -23.60 |
| Consensus MFE | -16.46 |
| Energy contribution | -16.55 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850439 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16664081 101 - 27905053 AAA----CAGAAAACUCACCACAAAUGAUGGCCAAUUCAUGGCCAUGUUAAUGCCAUGAUUGUUCGACCACCAUUUUAGGUCUUUCUGCUGGUCUGCAUUGUUGA .((----((((((....(((..(((((.((((.((((((((((.((....)))))))))..))).).))).)))))..))).))))(((......))).)))).. ( -23.60) >DroVir_CAF1 7510 104 - 1 UUUCCGACU-AAGACUUACCACAAAUGCUGGCCAAUUCAUGGCCAUAUUUAUGCCAUGAUUAUUUGACCACCAUUUCAAGUCCUUUUGUUGGUCCGCAUUGUUAA ...(((((.-..(((((.....(((((.(((.(((.(((((((.........)))))))....))).))).))))).))))).....)))))............. ( -25.90) >DroGri_CAF1 8053 98 - 1 UGUCU-------CUCUUACCACAAAUGCCGGCCAGUUCAUGGCCAUGUUAAUGCCAUGAUUAUUUGACCACCAUUUCAAGUCCUUUUGUUGGUCCGCAUUGUUAA .....-------...........(((((.((((((.(((((((.((....)))))))))..((((((........)))))).......)))))).)))))..... ( -25.00) >DroMoj_CAF1 6845 104 - 1 UA-CCAUCUAAACACUUACCACAAAUGCAGGCCAGUUCAUGGCCAUAUUGAUACCAUGGUUAUUUGACCACCAUUUCAAGUCCUUUUGUUGGUCCGCAUUGUUGA ..-................((..(((((.(((((.....))))).............((((....))))((((...((((....)))).))))..)))))..)). ( -23.10) >DroAna_CAF1 6320 100 - 1 AGU----CAGA-AACUCACCACAAAUGCCGGCCAGUUCAUAGCCAUGUUAAUACCAUAUUUAUUUGACCAAAAUUUGAGGUCUUUCUGCUGGUCCGCAUUGUUGA ...----....-.......((..(((((.(((((((....((((..(((((............)))))((.....)).)).))....))))))).)))))..)). ( -21.00) >DroPer_CAF1 8061 101 - 1 GGG----CACACAGCUUACCACAAAGAUUGGCCAGUUCAUGGCCAUGUUAAUACCAUGAUUGUUUGACCACCACUUCAGAUCCUUCUGCUGGUCCGCAUUGUUGA (((----(...((((.............((((((.....))))))............((..((((((........))))))...)).)))))))).......... ( -23.00) >consensus UGU____CA_AAAACUUACCACAAAUGCUGGCCAGUUCAUGGCCAUGUUAAUACCAUGAUUAUUUGACCACCAUUUCAAGUCCUUCUGCUGGUCCGCAUUGUUGA .......................(((((.((((((.((((((...........)))))).................((((....)))))))))).)))))..... (-16.46 = -16.55 + 0.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:19:36 2006