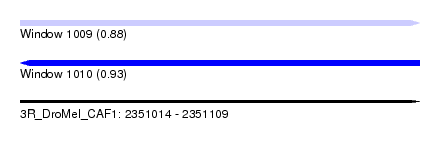
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,351,014 – 2,351,109 |
| Length | 95 |
| Max. P | 0.932632 |

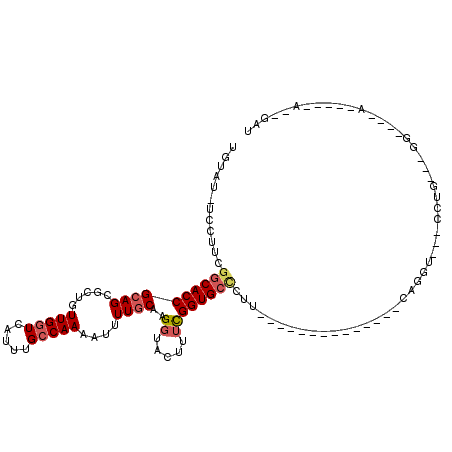
| Location | 2,351,014 – 2,351,109 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 67.19 |
| Mean single sequence MFE | -25.16 |
| Consensus MFE | -9.86 |
| Energy contribution | -10.70 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.39 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.876313 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2351014 95 + 27905053 AUCUAU----GUUAAGCC---CAAGAACACCUG--------------AAGGGCACCGAAAGUACCUUGCAAAACUUUGGCAAAUGACCAACAGCGCUGCGGUGCCGAAGGA-AUACA ......----.....(((---(.((.....)).--------------..)))).((....(((((..(((...(((((((....).)))).))...))))))))....)).-..... ( -21.60) >DroSec_CAF1 54806 95 + 1 AUCUAU----CUUAUUCC---CAGGAGCAUCUG--------------AAGGGCACCGAAACUACCUUGCAAAACUUUGGCAAAUGACCAACAGCGCUGCGGUGUCGCAGGA-AUACA ......----..((((((---(((......)))--------------...(((((((.......)..(((...(((((((....).)))).))...)))))))))...)))-))).. ( -20.60) >DroSim_CAF1 55165 116 + 1 AUCCGUUGAUUUGGUGCCCUUCAGGUGUUCUUGGUCAUAAGAUAGAUAAGGGCACCAAAAGUACCUUGCAAAAUUUUGGCAAUUGACCAACAGCGCUGCGGUGCCGCAGGA-AUACA ...(((((.((((((((((((((((....))))(((....)))....)))))))))))).((...((((.........))))...))...)))))(((((....)))))..-..... ( -39.10) >DroEre_CAF1 57479 79 + 1 ---------------------AACC---ACCUU--------------AAGUGCACCGAAAGUAACUUGCAAAAUGUUGGCAAAUGACCAACAGUGCUGCGGUGCCGAAGGAAAUACA ---------------------....---.((((--------------....((((((..........(((...(((((((....).)))))).)))..))))))..))))....... ( -24.80) >DroYak_CAF1 51186 79 + 1 ---------------------CAAG---ACCUU--------------UACAGCACCGAAUGUACCUUGCAAAAUGUUGCCAAAAGACCAAUUGCACUUCGGUGCCUAAGGAAAUACA ---------------------....---.((((--------------....((((((((.((.....((((...(((.......)))...))))))))))))))..))))....... ( -19.70) >consensus AUC__U_____U____CC___CAAG___ACCUG______________AAGGGCACCGAAAGUACCUUGCAAAAUUUUGGCAAAUGACCAACAGCGCUGCGGUGCCGAAGGA_AUACA ..................................................(((((((...((.....))......(((((....).))))........)))))))............ ( -9.86 = -10.70 + 0.84)

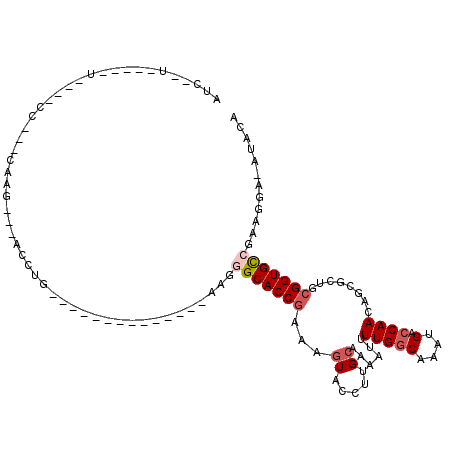
| Location | 2,351,014 – 2,351,109 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 67.19 |
| Mean single sequence MFE | -28.24 |
| Consensus MFE | -12.78 |
| Energy contribution | -13.50 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.45 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932632 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2351014 95 - 27905053 UGUAU-UCCUUCGGCACCGCAGCGCUGUUGGUCAUUUGCCAAAGUUUUGCAAGGUACUUUCGGUGCCCUU--------------CAGGUGUUCUUG---GGCUUAAC----AUAGAU ((((.-.(((..((((((((((.(((.(((((.....)))))))).))))..(((((.....)))))...--------------..))))))...)---))..).))----)..... ( -28.90) >DroSec_CAF1 54806 95 - 1 UGUAU-UCCUGCGACACCGCAGCGCUGUUGGUCAUUUGCCAAAGUUUUGCAAGGUAGUUUCGGUGCCCUU--------------CAGAUGCUCCUG---GGAAUAAG----AUAGAU ..(((-(((((.((((((((((.(((.(((((.....)))))))).))))..))).))).)((.((....--------------.....)).)).)---))))))..----...... ( -24.70) >DroSim_CAF1 55165 116 - 1 UGUAU-UCCUGCGGCACCGCAGCGCUGUUGGUCAAUUGCCAAAAUUUUGCAAGGUACUUUUGGUGCCCUUAUCUAUCUUAUGACCAAGAACACCUGAAGGGCACCAAAUCAACGGAU .....-..(((((....)))))..((((((((....((((............))))...(((((((((((.....((((......)))).......))))))))))))))))))).. ( -38.40) >DroEre_CAF1 57479 79 - 1 UGUAUUUCCUUCGGCACCGCAGCACUGUUGGUCAUUUGCCAACAUUUUGCAAGUUACUUUCGGUGCACUU--------------AAGGU---GGUU--------------------- ..............((((...((((((..(((.((((((.........)))))).)))..))))))....--------------..)))---)...--------------------- ( -22.60) >DroYak_CAF1 51186 79 - 1 UGUAUUUCCUUAGGCACCGAAGUGCAAUUGGUCUUUUGGCAACAUUUUGCAAGGUACAUUCGGUGCUGUA--------------AAGGU---CUUG--------------------- .......((((.(((((((((((((.....((....((....))....))...)))).)))))))))...--------------)))).---....--------------------- ( -26.60) >consensus UGUAU_UCCUUCGGCACCGCAGCGCUGUUGGUCAUUUGCCAAAAUUUUGCAAGGUACUUUCGGUGCCCUU______________CAGGU___CCUG___GG____A_____A__GAU ............((((((((((.....(((((.....)))))....))))..((.....)))))))).................................................. (-12.78 = -13.50 + 0.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:02 2006