
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,648,311 – 16,648,476 |
| Length | 165 |
| Max. P | 0.972765 |

| Location | 16,648,311 – 16,648,416 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 79.31 |
| Mean single sequence MFE | -28.18 |
| Consensus MFE | -20.44 |
| Energy contribution | -21.00 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.619457 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16648311 105 + 27905053 CAACUC------AGGGAUUUAGAGGUUGCAUUUACCUGGAGGCUUUGCGUUCCC-CCAGACGCUUAGCCUAGUCAUUAAUAAAUGCUAGAUA-AAGUGCCAGAAUCGGACAUU ....((------.((.(((((.((((.......))))..(((((..(((((...-...)))))..))))).........))))).)).))..-.((((((......)).)))) ( -26.10) >DroPse_CAF1 795 112 + 1 CAUAGCACAAUAUGUGCUUUCGCAUCCGCAUUUACCUGGAGGCUUUGUGUUCCG-CCAGACGCUUAGCCUAGUCUUUAAUAAAUGCCAUAGAAAAAUGCUUGAAUCGGACAUU ...(((((.....)))))......((((((((((...(((((((..(((((...-...)))))..))))...)))....)))))))(((......)))........))).... ( -27.10) >DroEre_CAF1 873 105 + 1 CAUCUC------GAGGAUUCCGAAGUUGCAUUUACCUGGAGGCUUUGCGUGACC-CCAGACGCUUAGCCUAGUCAUAAAUAAAUGCCAGAUA-AAGCGCCUGAGUCGGACAUU ......------......(((((((.(((.((((.(((((((((..((((....-....))))..)))))...(((......))))))).))-))))).))...))))).... ( -29.90) >DroYak_CAF1 859 105 + 1 CACCUC------AGGGAUUCUAAUGUUGCAUUUACCUGGAGGCUUUGCGUGACC-CCAGACGCUUAGCCUAGUCAUUUAUAAAUGCCAGAUA-AAGUGCCUGAAUCGGACAUU ..((((------(((.(((...((.(.(((((((..((((((((..((((....-....))))..)))))..)))....))))))).).)).-.))).)))))...))..... ( -29.60) >DroAna_CAF1 7523 105 + 1 CUUUCC------A-GCUUCCUGAAUUUGCAUUUACCUGGAGGCUUCGCGUGCCUGCAAGACGCCUAGCCUAGUCAUUAAUAAAUGCUAGAGA-AAAUGCCUGAAUCGGACAUU ......------.-.....((((.((.((((((.((((((((((..((((.........))))..)))))...((((....)))))))).).-))))))..)).))))..... ( -25.40) >DroPer_CAF1 812 112 + 1 CACAGCACAAUAUGUGCUUUCGAGUCCGCAUUUACCUGGAGGCUUUGUGUUCCG-CCAGACGCUUAGCCUAGUCUUUAAUAAAUGCCAUAGAAAAAUGCUUGAAUCGGACAUU ...(((((.....))))).....(((((((((((...(((((((..(((((...-...)))))..))))...)))....)))))))(((......)))........))))... ( -31.00) >consensus CAUCUC______AGGGAUUCCGAAGUUGCAUUUACCUGGAGGCUUUGCGUGCCC_CCAGACGCUUAGCCUAGUCAUUAAUAAAUGCCAGAGA_AAAUGCCUGAAUCGGACAUU ..................(((((....((((((..(((((((((..((((.........))))..)))))...((((....))))))))....)))))).....))))).... (-20.44 = -21.00 + 0.56)



| Location | 16,648,341 – 16,648,448 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 75.81 |
| Mean single sequence MFE | -29.25 |
| Consensus MFE | -25.44 |
| Energy contribution | -23.20 |
| Covariance contribution | -2.24 |
| Combinations/Pair | 1.61 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16648341 107 + 27905053 UGGAGGCUUUGCGUUCC-C-CCAGACGCUUAGCCUAGUCAUUAAUAAAUGCUAGAUA-AAGUGCCAGAAUCGGACAUUAUUCCCGCACUGAUCG-ACUGCAGAGCUGGAAU ...(((((..(((((..-.-...)))))..)))))((.((((....)))))).....-.((((((......)).))))(((((.((.(((....-....))).)).))))) ( -29.90) >DroVir_CAF1 1170 107 + 1 UGGAGGUUUUGUGUGCCUG-CUUGCCACUUAGCCUAGUCAAUAUUAAAUGCUAGAUC-AAAUGCUUGAGUUGGACAUUAUUUUCACAUUGAG--AUAAACAGUGUGAAGAU ...(((((..(((.((...-...)))))..)))))...........((((((((.((-((....)))).)))).))))((((((((((((..--.....)))))))))))) ( -29.60) >DroPse_CAF1 831 108 + 1 UGGAGGCUUUGUGUUCC-G-CCAGACGCUUAGCCUAGUCUUUAAUAAAUGCCAUAGAAAAAUGCUUGAAUCGGACAUUAUUCCCACAUUGACUG-CCAACAGUGCGGGAAU ...(((((..(((((..-.-...)))))..))))).((((.........((.((......)))).......))))...((((((.(((((....-....))))).)))))) ( -28.69) >DroMoj_CAF1 1178 107 + 1 UGGAGGCUUCCUGCGCCUG-CUUGCCGCUUAGCCUAGUCAUUAAUAAAUGCCAGAUC-AAAUGCAUAAAUUGGACAUUAUUUUCACACUGAA--ACAAUUAGUGUAAAGAU ...(((((....(((.(..-...).)))..))))).(((..((((..((((......-....))))..)))))))...(((((.(((((((.--....))))))).))))) ( -22.80) >DroAna_CAF1 7552 107 + 1 UGGAGGCUUCGCGUGCC-UGCAAGACGCCUAGCCUAGUCAUUAAUAAAUGCUAGAGA-AAAUGCCUGAAUCGGACAUUACUUUGGCCCUGAUU--CUCCCAGGGUCAGAGU ...((((.((...((..-..)).)).))))...((((.((((....))))))))...-.((((((......)).))))((((((((((((...--....)))))))))))) ( -35.80) >DroPer_CAF1 848 108 + 1 UGGAGGCUUUGUGUUCC-G-CCAGACGCUUAGCCUAGUCUUUAAUAAAUGCCAUAGAAAAAUGCUUGAAUCGGACAUUAUUCCCACAUUGACUG-CCAACAGUGCGGGAAU ...(((((..(((((..-.-...)))))..))))).((((.........((.((......)))).......))))...((((((.(((((....-....))))).)))))) ( -28.69) >consensus UGGAGGCUUUGUGUGCC_G_CCAGACGCUUAGCCUAGUCAUUAAUAAAUGCCAGAGA_AAAUGCUUGAAUCGGACAUUAUUCCCACACUGACU__CCAACAGUGCGAGAAU ...(((((..(((.((.......)))))..)))))........................((((((......)).))))((((((((((((.........)))))))))))) (-25.44 = -23.20 + -2.24)

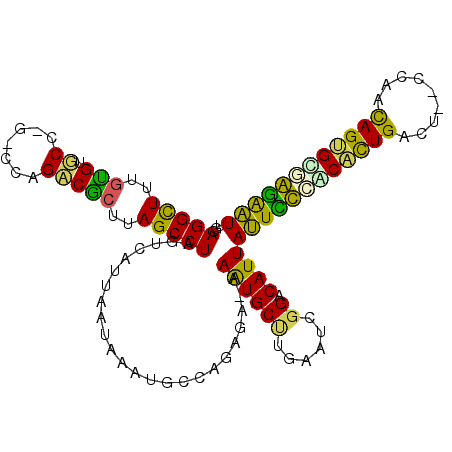

| Location | 16,648,378 – 16,648,476 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 80.85 |
| Mean single sequence MFE | -30.73 |
| Consensus MFE | -26.69 |
| Energy contribution | -26.08 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.13 |
| Mean z-score | -4.39 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969637 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16648378 98 + 27905053 AUUAAUAAAUGCUAGAUA-AAGUGCCAGAAUCGGACAUUAUUCCCGCACUGAUCGACUGCAGAGCUGGAAUAUCACAUGGUUCAGGCUACAAUACC-CAU ..................-..(((((.((((((.....((((((.((.(((........))).)).)))))).....)))))).))).))......-... ( -25.00) >DroPse_CAF1 868 100 + 1 UUUAAUAAAUGCCAUAGAAAAAUGCUUGAAUCGGACAUUAUUCCCACAUUGACUGCCAACAGUGCGGGAAUAUUAUAUGGUUCAAGCGAUAAUUGACUAA ......................(((((((((((.....(((((((.(((((........))))).))))))).....)))))))))))............ ( -30.40) >DroSec_CAF1 922 96 + 1 AUUAAUAAAUGCUAGAUA-AAGUGCCUGAAUCGGACAUUAUUCCCGCACUGAUUGACUCCAGAGCAGGAAUAUCACAUGGUUCAGGCUACAAUAAA-A-- ..................-..((((((((((((.....((((((.((.(((........))).)).)))))).....)))))))))).))......-.-- ( -30.50) >DroSim_CAF1 923 98 + 1 AUUAAUAAAUGCUAGAUA-AAGUGCCUGAAUCGGACAUUAUUCCCGCACUGAUUGACUCCAGAGCAGGAAUAUCACAUGGUUCAGGCUACAGUACC-CGU ..................-..((((((((((((.....((((((.((.(((........))).)).)))))).....)))))))))).))......-... ( -30.50) >DroYak_CAF1 926 96 + 1 AUUUAUAAAUGCCAGAUA-AAGUGCCUGAAUCGGACAUUAUUCCUGCGCUGAUU-ACUUCAGAGCAGGAAUAUCAAAUGGUUCAGGCUACAAAUCC--CU ..................-..((((((((((((.....(((((((((.((((..-...)))).))))))))).....)))))))))).))......--.. ( -37.60) >DroPer_CAF1 885 100 + 1 UUUAAUAAAUGCCAUAGAAAAAUGCUUGAAUCGGACAUUAUUCCCACAUUGACUGCCAACAGUGCGGGAAUAUUAUAUGGUUCAAGCGAUAAUUGACUAA ......................(((((((((((.....(((((((.(((((........))))).))))))).....)))))))))))............ ( -30.40) >consensus AUUAAUAAAUGCCAGAUA_AAGUGCCUGAAUCGGACAUUAUUCCCGCACUGAUUGACUACAGAGCAGGAAUAUCACAUGGUUCAGGCUACAAUACA_CAU .....................((((((((((((.....((((((.((.(((........))).)).)))))).....)))))))))).)).......... (-26.69 = -26.08 + -0.61)

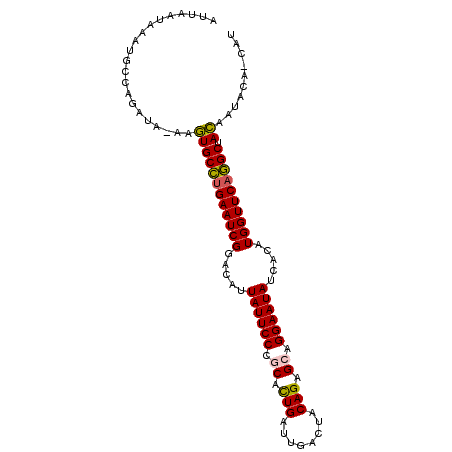
| Location | 16,648,378 – 16,648,476 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 80.85 |
| Mean single sequence MFE | -28.80 |
| Consensus MFE | -25.05 |
| Energy contribution | -24.77 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972765 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16648378 98 - 27905053 AUG-GGUAUUGUAGCCUGAACCAUGUGAUAUUCCAGCUCUGCAGUCGAUCAGUGCGGGAAUAAUGUCCGAUUCUGGCACUU-UAUCUAGCAUUUAUUAAU .((-((((..((.(((.(((........((((((.((.(((........))).)).))))))........))).)))))..-))))))............ ( -24.89) >DroPse_CAF1 868 100 - 1 UUAGUCAAUUAUCGCUUGAACCAUAUAAUAUUCCCGCACUGUUGGCAGUCAAUGUGGGAAUAAUGUCCGAUUCAAGCAUUUUUCUAUGGCAUUUAUUAAA ...((((......(((((((.(..(((.(((((((((((((....))))....))))))))).)))..).))))))).........)))).......... ( -27.26) >DroSec_CAF1 922 96 - 1 --U-UUUAUUGUAGCCUGAACCAUGUGAUAUUCCUGCUCUGGAGUCAAUCAGUGCGGGAAUAAUGUCCGAUUCAGGCACUU-UAUCUAGCAUUUAUUAAU --.-..((..((.(((((((........(((((((((.((((......)))).)))))))))........)))))))))..-))................ ( -28.09) >DroSim_CAF1 923 98 - 1 ACG-GGUACUGUAGCCUGAACCAUGUGAUAUUCCUGCUCUGGAGUCAAUCAGUGCGGGAAUAAUGUCCGAUUCAGGCACUU-UAUCUAGCAUUUAUUAAU ..(-((((..((.(((((((........(((((((((.((((......)))).)))))))))........)))))))))..-)))))............. ( -32.49) >DroYak_CAF1 926 96 - 1 AG--GGAUUUGUAGCCUGAACCAUUUGAUAUUCCUGCUCUGAAGU-AAUCAGCGCAGGAAUAAUGUCCGAUUCAGGCACUU-UAUCUGGCAUUUAUAAAU ..--((((..((.(((((((........(((((((((.((((...-..)))).)))))))))........)))))))))..-.))))............. ( -32.79) >DroPer_CAF1 885 100 - 1 UUAGUCAAUUAUCGCUUGAACCAUAUAAUAUUCCCGCACUGUUGGCAGUCAAUGUGGGAAUAAUGUCCGAUUCAAGCAUUUUUCUAUGGCAUUUAUUAAA ...((((......(((((((.(..(((.(((((((((((((....))))....))))))))).)))..).))))))).........)))).......... ( -27.26) >consensus AUA_GGAAUUGUAGCCUGAACCAUGUGAUAUUCCUGCUCUGGAGUCAAUCAGUGCGGGAAUAAUGUCCGAUUCAGGCACUU_UAUCUAGCAUUUAUUAAU .............(((((((........(((((((((.(((........))).)))))))))........)))))))....................... (-25.05 = -24.77 + -0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:19:28 2006