| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,592,816 – 16,592,963 |
| Length | 147 |
| Max. P | 0.986140 |

| Location | 16,592,816 – 16,592,925 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 79.53 |
| Mean single sequence MFE | -27.99 |
| Consensus MFE | -17.46 |
| Energy contribution | -18.63 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.762066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16592816 109 - 27905053 GUCCUAGAGUCUAA----UAUAAAAGUAGGUAUUAUUUUUGUAGCCAUGUAAGGCUAGAAAAAUGUUGCCACUUAGUCCUCUGCGGCAGGUA-ACGUGAAAACUGUUUGCCAAC (((.(((((.(((.----......(((.((((.(((((((.(((((......))))).))))))).))))))))))..))))).))).((((-((((....)).).)))))... ( -34.41) >DroSec_CAF1 149077 109 - 1 GUCCUACAAUCUAA----AAUAAAAGUAGGUAAUAUUUUUGUAGCCAUGUAAGGCUAGGAAAAUGUUGCCACUUAGUCCGCUGCGGCAGGUA-ACGUGAAAACUGUUUGCCAAC ..............----.....((((.((((((((((((.(((((......))))).))))))))))))))))..........((((..((-..((....))))..))))... ( -34.90) >DroSim_CAF1 154075 109 - 1 GUGCAAGAAUCUAA----UAUAAAAGUAGGUAUUAAUUUUGUAGCCAUGUAAGGCUAGGAAAAUGUUGCCACUUAGUCCUCUGCGGCAGGUA-ACGUGAAAACUGUUUGCCAAC ..(((.((..(((.----......(((.((((.((.((((.(((((......))))).)))).)).))))))))))...)))))((((..((-..((....))))..))))... ( -29.21) >DroEre_CAF1 152919 104 - 1 AUUCUAGAUCCUAA------UAAAUGGGUGUCUUAU-UUUGUAGCCAUG-CAGGCUAGGAAAAUGUUGCCACUUAGUCCUCUGCGGCAGGU--ACGUGAAAACUGUUUGCCAAC .....((((((((.------....)))).))))..(-(((.(((((...-..))))).))))......................((((..(--(.((....))))..))))... ( -25.30) >DroYak_CAF1 148561 107 - 1 GUCCUAGAUCCUUA----UAUAAAAGGAAGUGUUAU-UUUGUAGCCAUG-AAGGCUAGGAAAAUAUUGCCACUUAGUCCUCUGCGGCAGGUA-ACGUGAAAACUGUUUGCCAAC (((.(((((((((.----.....)))))((((.(((-(((.(((((...-..))))).))))....)).))))......)))).))).((((-((((....)).).)))))... ( -27.30) >DroAna_CAF1 145627 105 - 1 CUUCCCGAU-UUAAGGACAACAAAAUCGUGUACCAUUUUU--CCCAAUG------UAGAAAAAUGUUUCUAGUUUGGAUUAAGCCCCAGCUAAACGUGAAAACUGUUUGCCAAC .........-....((..((((...((((((..(((((((--(......------..))))))))....(((((.((.......)).))))).))))))....))))..))... ( -16.80) >consensus GUCCUAGAUUCUAA____UAUAAAAGUAGGUAUUAUUUUUGUAGCCAUG_AAGGCUAGGAAAAUGUUGCCACUUAGUCCUCUGCGGCAGGUA_ACGUGAAAACUGUUUGCCAAC .......................(((...(((.(((((((.(((((......))))).))))))).)))..))).(((......))).((((.((((....)).)).))))... (-17.46 = -18.63 + 1.17)


| Location | 16,592,855 – 16,592,963 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 76.66 |
| Mean single sequence MFE | -14.82 |
| Consensus MFE | -9.84 |
| Energy contribution | -9.68 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569726 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16592855 108 + 27905053 AAGUGGCAACAUUUUUCUAGCCUUACAUGGCUACAAAAAUAAUACCUACUUUUAUAUUAGACUCUAGGACCAAAUUAUUAAUUUAAUUCUCAAA-AAAAAAAUUCAAUC- ((((((....((((((.(((((......))))).)))))).....))))))...........................................-..............- ( -14.00) >DroSec_CAF1 149116 106 + 1 AAGUGGCAACAUUUUCCUAGCCUUACAUGGCUACAAAAAUAUUACCUACUUUUAUUUUAGAUUGUAGGACAAAAUUAUAAAUUUAAUUCUCAAA-AA--AAAUUGAAUC- (((((....)))))...(((((......)))))...........(((((..((......))..)))))............((((((((......-..--.)))))))).- ( -18.40) >DroSim_CAF1 154114 106 + 1 AAGUGGCAACAUUUUCCUAGCCUUACAUGGCUACAAAAUUAAUACCUACUUUUAUAUUAGAUUCUUGCACAAAAUUAUAAAUUUUAUUCUCAAA-AA--AAAUUGAAUC- ((((((......(((..(((((......)))))..))).......))))))........(((((..............................-..--.....)))))- ( -12.21) >DroEre_CAF1 152957 97 + 1 AAGUGGCAACAUUUUCCUAGCCUG-CAUGGCUACAAA-AUAAGACACCCAUUUA--UUAGGAUCUAGAAUCUG---GUUUAUUUAAUUCUCACA-AC-----UUGCAUUU (((((....)))))...(((((..-...)))))..((-(((((....((.....--...))..((((...)))---))))))))..........-..-----........ ( -13.40) >DroYak_CAF1 148600 97 + 1 AAGUGGCAAUAUUUUCCUAGCCUU-CAUGGCUACAAA-AUAACACUUCCUUUUAUAUAAGGAUCUAGGACAAA------AAUUCGUAUCUCAUAUAC-----UUUCAUUU ((((((........((((((((..-...)))......-........(((((......)))))..)))))....------.....((((.....))))-----..)))))) ( -16.10) >consensus AAGUGGCAACAUUUUCCUAGCCUUACAUGGCUACAAAAAUAAUACCUACUUUUAUAUUAGAAUCUAGGACAAAAUUAUAAAUUUAAUUCUCAAA_AA__AAAUUGAAUC_ (((((....)))))...(((((......)))))............................................................................. ( -9.84 = -9.68 + -0.16)

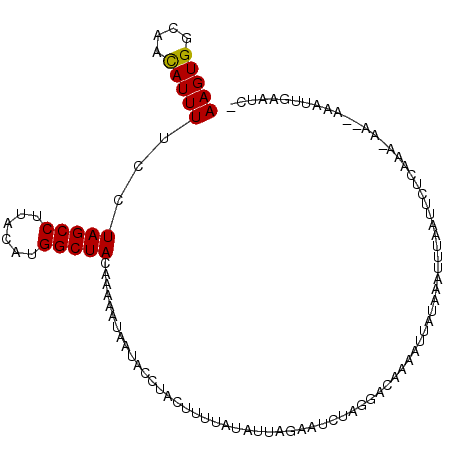
| Location | 16,592,855 – 16,592,963 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 76.66 |
| Mean single sequence MFE | -23.24 |
| Consensus MFE | -15.06 |
| Energy contribution | -15.82 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986140 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16592855 108 - 27905053 -GAUUGAAUUUUUUU-UUUGAGAAUUAAAUUAAUAAUUUGGUCCUAGAGUCUAAUAUAAAAGUAGGUAUUAUUUUUGUAGCCAUGUAAGGCUAGAAAAAUGUUGCCACUU -(((..((((.((..-(((((...)))))..)).))))..)))................((((.((((.(((((((.(((((......))))).))))))).)))))))) ( -27.60) >DroSec_CAF1 149116 106 - 1 -GAUUCAAUUU--UU-UUUGAGAAUUAAAUUUAUAAUUUUGUCCUACAAUCUAAAAUAAAAGUAGGUAAUAUUUUUGUAGCCAUGUAAGGCUAGGAAAAUGUUGCCACUU -(((((((...--..-.)))(((((((......))))))).......))))........((((.((((((((((((.(((((......))))).)))))))))))))))) ( -27.50) >DroSim_CAF1 154114 106 - 1 -GAUUCAAUUU--UU-UUUGAGAAUAAAAUUUAUAAUUUUGUGCAAGAAUCUAAUAUAAAAGUAGGUAUUAAUUUUGUAGCCAUGUAAGGCUAGGAAAAUGUUGCCACUU -..........--((-((((...(((((((.....))))))).))))))..........((((.((((.((.((((.(((((......))))).)))).)).)))))))) ( -23.20) >DroEre_CAF1 152957 97 - 1 AAAUGCAA-----GU-UGUGAGAAUUAAAUAAAC---CAGAUUCUAGAUCCUAA--UAAAUGGGUGUCUUAU-UUUGUAGCCAUG-CAGGCUAGGAAAAUGUUGCCACUU ....((((-----..-....((((((........---..))))))((((((((.--....)))).))))..(-(((.(((((...-..))))).))))...))))..... ( -19.70) >DroYak_CAF1 148600 97 - 1 AAAUGAAA-----GUAUAUGAGAUACGAAUU------UUUGUCCUAGAUCCUUAUAUAAAAGGAAGUGUUAU-UUUGUAGCCAUG-AAGGCUAGGAAAAUAUUGCCACUU ........-----.((((((((((((((...------.)))).....)).))))))))...((.((((((..-(((.(((((...-..))))).))))))))).)).... ( -18.20) >consensus _GAUUCAAUUU__UU_UUUGAGAAUUAAAUUAAUAAUUUUGUCCUAGAAUCUAAUAUAAAAGUAGGUAUUAUUUUUGUAGCCAUGUAAGGCUAGGAAAAUGUUGCCACUU ................................................................((((.(((((((.(((((......))))).))))))).)))).... (-15.06 = -15.82 + 0.76)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:19:09 2006