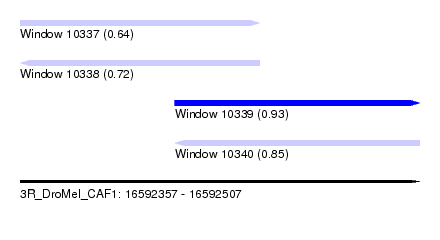
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,592,357 – 16,592,507 |
| Length | 150 |
| Max. P | 0.933096 |

| Location | 16,592,357 – 16,592,447 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 78.96 |
| Mean single sequence MFE | -22.88 |
| Consensus MFE | -13.44 |
| Energy contribution | -14.97 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.636023 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16592357 90 + 27905053 UAUACCCUUCCUCCCGAAUGCCAAUCAAGUCUGGCUGUCCUGCGAGCACUACAACUAGUUCUGCCAGCCGUCCUCGUCUUUAGAGGACGA ...............((..((((........))))..))..((..(((..((.....))..)))..))(((((((.......))))))). ( -23.90) >DroSec_CAF1 148615 90 + 1 CAUCCCCUUCCUCCCGAAUGCCAAUCAAGUCUGGCUGUCCUGCGAACACUACAACUAGUUCUGCCAGCCGUCCUCGUCCUCAGAGGACGA ................................(((((....((((((..........)))).)))))))((((((.......)))))).. ( -22.20) >DroSim_CAF1 153612 90 + 1 CAUCCCCUUCCUCCCGAAUGCCAAUCAAGUCUGGCUGUCCUGCGAACACUACAACUAGUUCUGCCAGCCGUCCUCGUCCUCAGAGGACGA ................................(((((....((((((..........)))).)))))))((((((.......)))))).. ( -22.20) >DroEre_CAF1 152451 85 + 1 UAUCCCCUCCC-----CAUGCCAAUCAAGUCUGGCUGUCCUGCGAGCACUACAACUAGUUCUGCCAGCCGUCCUCGUCCUCAGAGGACGA ...........-----((.((((........))))))....((..(((..((.....))..)))..))(((((((.......))))))). ( -23.50) >DroYak_CAF1 148099 85 + 1 UAUCCCCUCCC-----CAUGCCAAUCAAGUCUUGCUGUCCUGCGAGCACUACAACUAGUUUUGCCAGCCGUCCUCGUCCUCAGAGGACGA ...........-----.................((((....(((((.((((....)))))))))))))(((((((.......))))))). ( -21.70) >DroAna_CAF1 145166 70 + 1 GACAG---UCAGCCAGUCGGCCAGGCCAGGCAGCCUGUCCUGC-----CUACAACUAGUUCUGUCAGUC------------GGAGGACAA (((((---..(((.(((.(((.(((.((((...)))).)))))-----)....))).)))))))).(((------------....))).. ( -23.80) >consensus UAUCCCCUUCCUCCCGAAUGCCAAUCAAGUCUGGCUGUCCUGCGAGCACUACAACUAGUUCUGCCAGCCGUCCUCGUCCUCAGAGGACGA ................................(((((....(((...((((....))))..))))))))((((((.......)))))).. (-13.44 = -14.97 + 1.53)



| Location | 16,592,357 – 16,592,447 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 78.96 |
| Mean single sequence MFE | -28.78 |
| Consensus MFE | -21.51 |
| Energy contribution | -22.65 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.719562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16592357 90 - 27905053 UCGUCCUCUAAAGACGAGGACGGCUGGCAGAACUAGUUGUAGUGCUCGCAGGACAGCCAGACUUGAUUGGCAUUCGGGAGGAAGGGUAUA .(((((((.......)))))))(((((.....)))))....((((((........(((((......))))).(((....))).)))))). ( -28.40) >DroSec_CAF1 148615 90 - 1 UCGUCCUCUGAGGACGAGGACGGCUGGCAGAACUAGUUGUAGUGUUCGCAGGACAGCCAGACUUGAUUGGCAUUCGGGAGGAAGGGGAUG .((((((((..(((((...((((((((.....))))))))..)))))........(((((......))))).(((....))))))))))) ( -31.10) >DroSim_CAF1 153612 90 - 1 UCGUCCUCUGAGGACGAGGACGGCUGGCAGAACUAGUUGUAGUGUUCGCAGGACAGCCAGACUUGAUUGGCAUUCGGGAGGAAGGGGAUG .((((((((..(((((...((((((((.....))))))))..)))))........(((((......))))).(((....))))))))))) ( -31.10) >DroEre_CAF1 152451 85 - 1 UCGUCCUCUGAGGACGAGGACGGCUGGCAGAACUAGUUGUAGUGCUCGCAGGACAGCCAGACUUGAUUGGCAUG-----GGGAGGGGAUA ..(((((.((((.((....((((((((.....)))))))).)).)))).))))).(((((......)))))...-----........... ( -30.60) >DroYak_CAF1 148099 85 - 1 UCGUCCUCUGAGGACGAGGACGGCUGGCAAAACUAGUUGUAGUGCUCGCAGGACAGCAAGACUUGAUUGGCAUG-----GGGAGGGGAUA .(((((((.......)))))))((..((((..((.(((((..((....))..))))).))..))).)..))...-----........... ( -26.30) >DroAna_CAF1 145166 70 - 1 UUGUCCUCC------------GACUGACAGAACUAGUUGUAG-----GCAGGACAGGCUGCCUGGCCUGGCCGACUGGCUGA---CUGUC ..(((....------------))).(((((..(((((((..(-----.((((.((((...)))).)))).))))))))....---))))) ( -25.20) >consensus UCGUCCUCUGAGGACGAGGACGGCUGGCAGAACUAGUUGUAGUGCUCGCAGGACAGCCAGACUUGAUUGGCAUUCGGGAGGAAGGGGAUA ..(((((.((((.((....((((((((.....)))))))).)).)))).))))).(((((......)))))................... (-21.51 = -22.65 + 1.14)


| Location | 16,592,415 – 16,592,507 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 79.75 |
| Mean single sequence MFE | -27.70 |
| Consensus MFE | -15.10 |
| Energy contribution | -14.49 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933096 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16592415 92 + 27905053 UUCUGCCAGCCGUCCUCGUCUUUAGAGGACGACGACAGCAAAGGCAGUAAC--AGCGGCAGCAAAUAAUUUUGUAAUUCAAUUGUUGUCCUUCU ..(((((.((.(((.((((((.....)))))).))).))...)))))....--((.((((((((..((((....))))...))))))))))... ( -30.00) >DroPse_CAF1 150390 80 + 1 UUCUUCAAGUC------------AGACGAGGACGUGGACGUGGACGACGAC--AGCAGCAGCAAAUAAUUUUGUAAUUCAAUUGUUGUCGUUCU ........(((------------..(((....))).)))..((((((((((--((.....(((((....))))).......)))))))))))). ( -22.60) >DroSim_CAF1 153670 94 + 1 UUCUGCCAGCCGUCCUCGUCCUCAGAGGACGACGACAGCAAAGGCAGUAACACAGCGGCAGCAAAUAAUUUUGUAAUUCAAUUGUUGUCCUUCU ..(((((.((.(((.((((((.....)))))).))).))...)))))......((.((((((((..((((....))))...))))))))))... ( -32.30) >DroEre_CAF1 152504 92 + 1 UUCUGCCAGCCGUCCUCGUCCUCAGAGGACGACGACAGCAAAGGCAGUAAG--AGCAGCAGCAAAUAAUUUUGUAAUUCAAUUGUUGUCCUUCU ..(((((.((.(((.((((((.....)))))).))).))...))))).(((--..((((((((((....)))))........)))))..))).. ( -32.50) >DroYak_CAF1 148152 92 + 1 UUUUGCCAGCCGUCCUCGUCCUCAGAGGACGACGACAGCAGAAGCAGUAAC--AGCAGCAGCAAAUAAUUUUGUAAUUCAAUUGUUGUCCUUCU ........(.(((((((.......))))))).)((((((((..((.((...--.)).)).(((((....))))).......))))))))..... ( -26.20) >DroPer_CAF1 157725 80 + 1 UUCUUCAAGUC------------AGACGAGGACGAGGACGUGGACGACGAC--AGCAGCAGCAAAUAAUUUUGUAAUUCAAUUGUUGUCGUUCU .(((((..(((------------.......))))))))...((((((((((--((.....(((((....))))).......)))))))))))). ( -22.60) >consensus UUCUGCCAGCCGUCCUCGUCCUCAGAGGACGACGACAGCAAAGGCAGUAAC__AGCAGCAGCAAAUAAUUUUGUAAUUCAAUUGUUGUCCUUCU ........................(((((((((((........((((.......((....))........)))).......))))))))))).. (-15.10 = -14.49 + -0.61)



| Location | 16,592,415 – 16,592,507 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 79.75 |
| Mean single sequence MFE | -26.70 |
| Consensus MFE | -15.71 |
| Energy contribution | -18.63 |
| Covariance contribution | 2.92 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16592415 92 - 27905053 AGAAGGACAACAAUUGAAUUACAAAAUUAUUUGCUGCCGCU--GUUACUGCCUUUGCUGUCGUCGUCCUCUAAAGACGAGGACGGCUGGCAGAA ....((.((.(((...((((....))))..))).))))...--....(((((...((((((.(((((.......))))).)))))).))))).. ( -32.90) >DroPse_CAF1 150390 80 - 1 AGAACGACAACAAUUGAAUUACAAAAUUAUUUGCUGCUGCU--GUCGUCGUCCACGUCCACGUCCUCGUCU------------GACUUGAAGAA .(.(((((.(((.(((.....)))........((....)))--)).))))).)..(((.(((....)))..------------)))........ ( -12.70) >DroSim_CAF1 153670 94 - 1 AGAAGGACAACAAUUGAAUUACAAAAUUAUUUGCUGCCGCUGUGUUACUGCCUUUGCUGUCGUCGUCCUCUGAGGACGAGGACGGCUGGCAGAA ....((.((.(((...((((....))))..))).)))).........(((((...((((((.((((((.....)))))).)))))).))))).. ( -37.20) >DroEre_CAF1 152504 92 - 1 AGAAGGACAACAAUUGAAUUACAAAAUUAUUUGCUGCUGCU--CUUACUGCCUUUGCUGUCGUCGUCCUCUGAGGACGAGGACGGCUGGCAGAA (((.((.((.(((...((((....))))..))).))))..)--))..(((((...((((((.((((((.....)))))).)))))).))))).. ( -36.10) >DroYak_CAF1 148152 92 - 1 AGAAGGACAACAAUUGAAUUACAAAAUUAUUUGCUGCUGCU--GUUACUGCUUCUGCUGUCGUCGUCCUCUGAGGACGAGGACGGCUGGCAAAA ....((.((.(((...((((....))))..))).))))...--.....((((...((((((.((((((.....)))))).)))))).))))... ( -29.40) >DroPer_CAF1 157725 80 - 1 AGAACGACAACAAUUGAAUUACAAAAUUAUUUGCUGCUGCU--GUCGUCGUCCACGUCCUCGUCCUCGUCU------------GACUUGAAGAA .(.(((((.(((.(((.....)))........((....)))--)).))))).)..(((..((....))...------------)))........ ( -11.90) >consensus AGAAGGACAACAAUUGAAUUACAAAAUUAUUUGCUGCUGCU__GUUACUGCCUUUGCUGUCGUCGUCCUCUGAGGACGAGGACGGCUGGCAGAA ....((.((.(((...((((....))))..))).)))).........(((((...((((((.(((((.......))))).)))))).))))).. (-15.71 = -18.63 + 2.92)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:19:06 2006