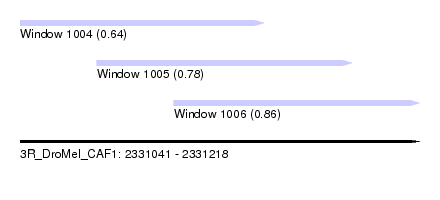
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,331,041 – 2,331,218 |
| Length | 177 |
| Max. P | 0.864886 |

| Location | 2,331,041 – 2,331,149 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.58 |
| Mean single sequence MFE | -32.67 |
| Consensus MFE | -20.20 |
| Energy contribution | -20.23 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2331041 108 + 27905053 GCUUUGUGAGAGGGCUU------GUCAGUUGGCUUGUAGAUUGAAAGCA------AACGCAAAUUGAAGAACAACUUCAGCAUGUGCACAUGCAGAGCUUAAUGAGAUGAGUUCUGCCAA ....((((.....((((------.((((((........)))))))))).------..((((..((((((.....))))))..)))))))).((((((((((......))))))))))... ( -32.80) >DroPse_CAF1 40345 120 + 1 GCUUUGUCAGUGGGCAUGGGCAUGUACAUAUGUAUGUAGAUCGAAAGCAGAGGCAGACGCAAAUUGAAGAACAACUUCAGCAUGUGCACAUGCAGAGCUUAAUGAGAUGAGUUCUGCCAA ((((((((....)))).))))((((.(((((((.(((....(....).....))).........(((((.....)))))))))))).))))((((((((((......))))))))))... ( -36.30) >DroEre_CAF1 42136 106 + 1 GCUUUGUGCU--GGCUU------GUCAGUUGGCUUGUAGAUUGAAAGCA------AACGCAAAUUGAAGAACAACUUCAGCAUGUGCACAUGCAGAGCUUAAUGAGAUGAGUUCUGCCAA ....(((((.--.((((------.((((((........)))))))))).------...(((..((((((.....))))))..)))))))).((((((((((......))))))))))... ( -34.30) >DroYak_CAF1 36606 106 + 1 ACGUUGUGUG--AGCUU------CUCAGUUGGCUUGUAGAUUGAAAGCA------AAUGCAAAUUGAAGAACAACUUCAGCAUGUGCACAUGCAGAGCUUAAUGAGAUGAGUUCUGCCAA ....(((((.--.((((------.((((((........)))))))))).------...(((..((((((.....))))))..)))))))).((((((((((......))))))))))... ( -32.70) >DroAna_CAF1 37916 106 + 1 UCAUUGUUCG--AGCUU------GUUAGCUGGCUUGUAGAUUGAAACCA------AACGCAAAUUGAAGAACAACUUCAGAGUGUGCACAUGCAAAGCUUAAUGAGAUGAGUUCUGCCAA (((.....((--((((.------(....).)))))).....))).....------..((((..((((((.....))))))..)))).....(((.((((((......)))))).)))... ( -24.80) >DroPer_CAF1 40197 116 + 1 GCUUUGUCAGUGGGCAUGGGCAUGUACAUAU----GUAGAUAGAAAGCAGAGGCAGACGCAAAUUGAAGAACAACUUCAGCAUGUGCACAUGCAGAGCUUAAUGAGAUGAGUUCUGCCAA ((((((((....)))).))))((((.(((((----((.........((....))..........(((((.....)))))))))))).))))((((((((((......))))))))))... ( -35.10) >consensus GCUUUGUGAG__GGCUU______GUCAGUUGGCUUGUAGAUUGAAAGCA______AACGCAAAUUGAAGAACAACUUCAGCAUGUGCACAUGCAGAGCUUAAUGAGAUGAGUUCUGCCAA .........................................................((((..((((((.....))))))..)))).....((((((((((......))))))))))... (-20.20 = -20.23 + 0.03)
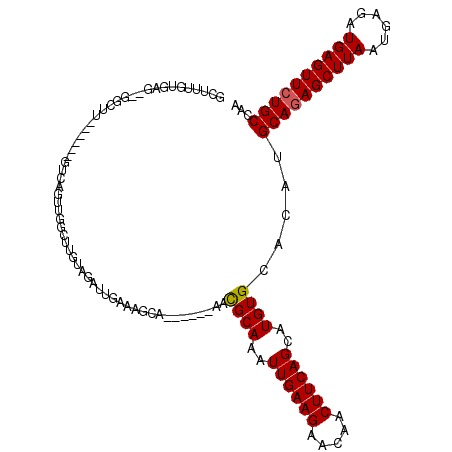
| Location | 2,331,075 – 2,331,188 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.46 |
| Mean single sequence MFE | -33.77 |
| Consensus MFE | -27.55 |
| Energy contribution | -27.67 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775104 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2331075 113 + 27905053 UUGAAAGCA------AACGCAAAUUGAAGAACAACUUCAGCAUGUGCACAUGCAGAGCUUAAUGAGAUGAGUUCUGCCAAGUGGCGUGUGUAAUGUGACAGGG-GCGUGGGGUGCACAGG ......(((------.((((....(((((.....)))))(((..(((.(((((((((((((......))))))))))...))))))..)))............-))))....)))..... ( -37.20) >DroPse_CAF1 40385 117 + 1 UCGAAAGCAGAGGCAGACGCAAAUUGAAGAACAACUUCAGCAUGUGCACAUGCAGAGCUUAAUGAGAUGAGUUCUGCCAAGUGGCGUGUGUAAUGUGGCAGGGGGCAUGGG---CAUGAU ......((....((....))....(((((.....)))))))..((((.(((((((((((((......)))))))).((..((.((((.....)))).))..)).))))).)---)))... ( -35.70) >DroEre_CAF1 42168 110 + 1 UUGAAAGCA------AACGCAAAUUGAAGAACAACUUCAGCAUGUGCACAUGCAGAGCUUAAUGAGAUGAGUUCUGCCAAGUGGUGUGUGUAAUGUGACAGGG-ACGUGGG---CGCAGU (((...((.------.((((....(((((.....)))))(((((..(((((((((((((((......))))))))))......)))))..).))))......)-.)))..)---).))). ( -32.70) >DroYak_CAF1 36638 110 + 1 UUGAAAGCA------AAUGCAAAUUGAAGAACAACUUCAGCAUGUGCACAUGCAGAGCUUAAUGAGAUGAGUUCUGCCAAGUGGGGUGUGUAAUGUGACAGGG-GCGUGGG---UGCAGU ......(((------.((((....(((((.....)))))((((.(((((((((((((((((......))))))))))........))))))))))).......-))))...---)))... ( -34.10) >DroAna_CAF1 37948 100 + 1 UUGAAACCA------AACGCAAAUUGAAGAACAACUUCAGAGUGUGCACAUGCAAAGCUUAAUGAGAUGAGUUCUGCCAAGUGGCGUGUGUAAUGUGACGGGG--CGC------------ ......((.------..((((..((((((.....))))))....(((((((((...((((....(((.....)))...)))).))))))))).))))...)).--...------------ ( -27.20) >DroPer_CAF1 40233 117 + 1 UAGAAAGCAGAGGCAGACGCAAAUUGAAGAACAACUUCAGCAUGUGCACAUGCAGAGCUUAAUGAGAUGAGUUCUGCCAAGUGGCGUGUGUAAUGUGGCAGGGGGCAUGGG---CAUGAU ......((....((....))....(((((.....)))))))..((((.(((((((((((((......)))))))).((..((.((((.....)))).))..)).))))).)---)))... ( -35.70) >consensus UUGAAAGCA______AACGCAAAUUGAAGAACAACUUCAGCAUGUGCACAUGCAGAGCUUAAUGAGAUGAGUUCUGCCAAGUGGCGUGUGUAAUGUGACAGGG_GCGUGGG___CACAGU .................((((...(((((.....)))))(((..(((.(((((((((((((......))))))))))...))))))..)))..))))....................... (-27.55 = -27.67 + 0.11)

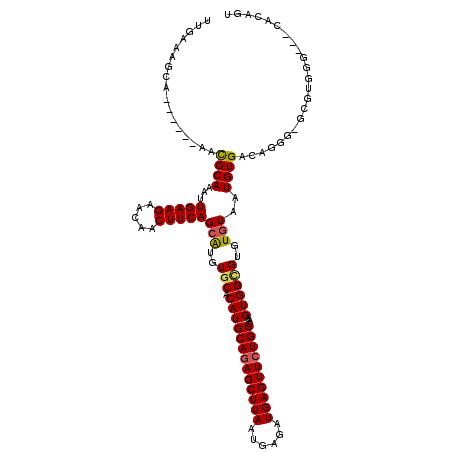
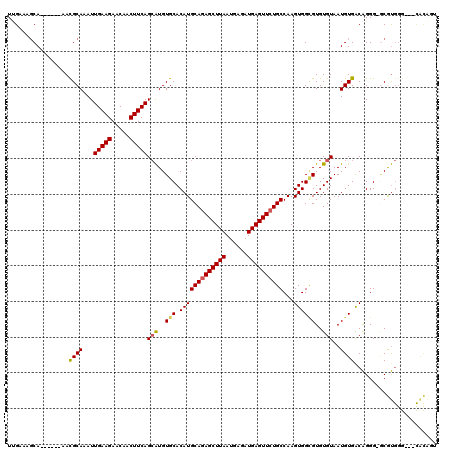
| Location | 2,331,109 – 2,331,218 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.46 |
| Mean single sequence MFE | -39.18 |
| Consensus MFE | -22.71 |
| Energy contribution | -22.35 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2331109 109 + 27905053 CAUGUGCACAUGCAGAGCUUAAUGAGAUGAGUUCUGCCAAGUGGCGUGUGUAAUGUGACAGGG-GCGUGGGGUGCACAGGGGCGGUACUGAUGGUA---------CUGAUG-GUACUGAA (((((.(....((((((((((......))))))))))...((.((((.....)))).))..).-))))).(((((.((....((((((.....)))---------))).))-)))))... ( -39.20) >DroSec_CAF1 36790 118 + 1 CAUGUGCACAUGCAGAGCUUAAUGAGAUGAGUUCUGCCAAGUGGCGUGUGUAAUGUGACAGGG-GCGUGGGGUGCGCCGGGGCGGUACUGAUGGUACUGAUGGUACUGAUG-GUACUGAA ((..(((.(((((((((((((......))))))))))...))))))..))............(-((((.....)))))....((((((((.((((((.....)))))).))-)))))).. ( -44.00) >DroSim_CAF1 37040 118 + 1 CAUGUGCACAUGCAGAGCUUAAUGAGAUGAGUUCUGCCAAGUGGCGUGUGUAAUGUGACAGGG-GCGUGGGGUGCGCCGGGGCGGUACUGAUGGUACUGAUGGUACUGAUG-GUACUGAA ((..(((.(((((((((((((......))))))))))...))))))..))............(-((((.....)))))....((((((((.((((((.....)))))).))-)))))).. ( -44.00) >DroEre_CAF1 42202 106 + 1 CAUGUGCACAUGCAGAGCUUAAUGAGAUGAGUUCUGCCAAGUGGUGUGUGUAAUGUGACAGGG-ACGUGGG---CGCAGUGGCGCUACUGACGGUA---------CUGAUG-GUACUGAU (((((.(....((((((((((......))))))))))...((.(..(.....)..).))..).-)))))((---(((....))))).....(((((---------(.....-)))))).. ( -36.80) >DroYak_CAF1 36672 100 + 1 CAUGUGCACAUGCAGAGCUUAAUGAGAUGAGUUCUGCCAAGUGGGGUGUGUAAUGUGACAGGG-GCGUGGG---UGCAGU------ACCGCCGGUA---------CUGAUG-CUACUAAA (((((((((((((((((((((......))))))))))........)))))).)))))..((..-((((.((---(((.(.------.....).)))---------)).)))-)..))... ( -34.20) >DroPer_CAF1 40273 104 + 1 CAUGUGCACAUGCAGAGCUUAAUGAGAUGAGUUCUGCCAAGUGGCGUGUGUAAUGUGGCAGGGGGCAUGGG---CAUGAU------GCUG------C-GAUGGCAAUGGCAUCCAGUCAA (((((.(....((((((((((......))))))))))...((.((((.....)))).))...).)))))((---(..(((------((((------(-....))...))))))..))).. ( -36.90) >consensus CAUGUGCACAUGCAGAGCUUAAUGAGAUGAGUUCUGCCAAGUGGCGUGUGUAAUGUGACAGGG_GCGUGGG___CGCAGGGGCGGUACUGAUGGUA_________CUGAUG_GUACUGAA (((((.(....((((((((((......))))))))))...((.((((.....)))).))...).)))))................................................... (-22.71 = -22.35 + -0.36)

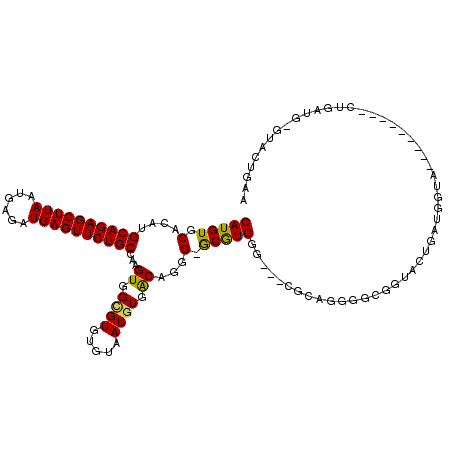
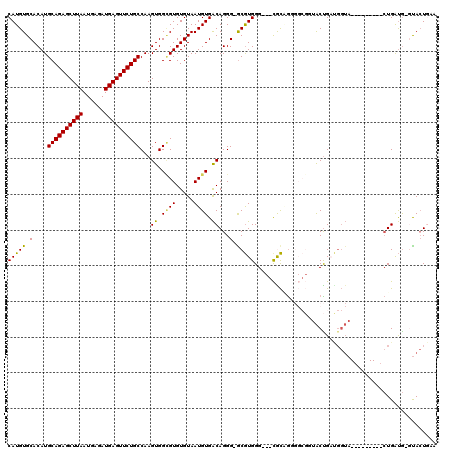
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:58 2006