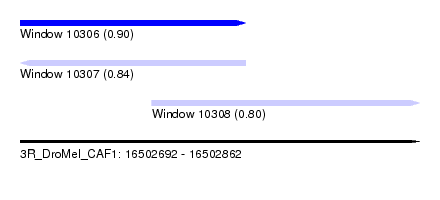
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,502,692 – 16,502,862 |
| Length | 170 |
| Max. P | 0.904800 |

| Location | 16,502,692 – 16,502,788 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 79.72 |
| Mean single sequence MFE | -37.05 |
| Consensus MFE | -25.68 |
| Energy contribution | -26.93 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
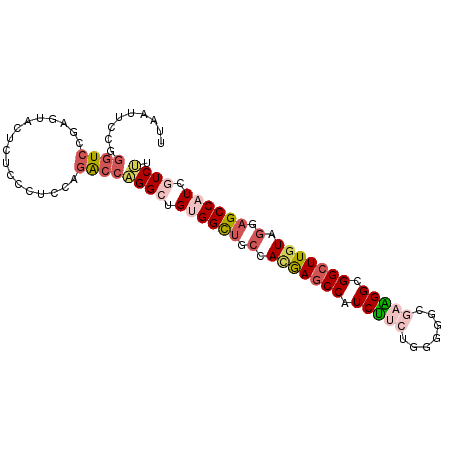
>3R_DroMel_CAF1 16502692 96 + 27905053 UUAGUUCCGGGUCCGAGUACUCUCCCUCCAGACCAGGCUGUGGCUGCCACGAGCCAUCUUCGGGGGGCGAAGGCGGCUUGUAGGAGCCAUCGUCUU .........((((.(((........)))..))))((((.((((((.(((((((((..(((((.....)))))..))))))).)))))))).)))). ( -42.60) >DroVir_CAF1 64266 89 + 1 -------GGACUCCACAUAUUCGCCCUCCAGGCCCGGCUGUGGUUUCCACAAACCAUCAUCUGCGUCAAUUGGCGGCUUAUAGCUGCCAUCAUCCU -------(((..(((((.....(((.....))).....)))))..)))......................(((((((.....)))))))....... ( -27.30) >DroSec_CAF1 58960 96 + 1 UUAAUUCCGGGUCCGAGUACUCUCCCUCCAGACCAGGCUGUGGCUGCCACGAGCCAUCUUCUGGUGGCGAAGGCGGCUUGUAGGAGCCAUCGUCUU .........((((.(((........)))..))))((((.((((((.(((((((((..((((.......))))..))))))).)))))))).)))). ( -39.90) >DroSim_CAF1 62986 96 + 1 UUAAUUCCGGGUCCGAGUACUCUCCCUCCAGACCAGGCUGUGGCUGCCACGAGCCAUCUUCUGGUGGCGAAGGCGGCUUGUAGGAGCCAUCGUCUU .........((((.(((........)))..))))((((.((((((.(((((((((..((((.......))))..))))))).)))))))).)))). ( -39.90) >DroEre_CAF1 60770 96 + 1 UUAAUUCAGGGUCGGAGUACUCUCCCUCCAGACCAGGCUGCGGCUGCCACGAGCCAUCUGCGGGGGGCAAAGGCGGCUUGUAGGAGCCAUCGUCUU .........((((((((........)))).))))((((...((((.(((((((((..((((.....))..))..))))))).))))))...)))). ( -36.90) >DroAna_CAF1 56438 96 + 1 UCAAUUCGGGGUCCACGUACUCUCCCUCCAAGCCCGGCUGGGGCUUCCAUAAGCCAUCCUCCGCACUCGAGGGUGGCUUGUACGAGCCAUCAUCGU .......(((((...(((((.........((((((.....))))))....(((((((((((.......)))))))))))))))).))).))..... ( -35.70) >consensus UUAAUUCCGGGUCCGAGUACUCUCCCUCCAGACCAGGCUGUGGCUGCCACGAGCCAUCUUCUGGGGGCGAAGGCGGCUUGUAGGAGCCAUCGUCUU .........((((.................))))((((.((((((.(.(((((((.(((((.......))))).))))))).).)))))).)))). (-25.68 = -26.93 + 1.25)

| Location | 16,502,692 – 16,502,788 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 79.72 |
| Mean single sequence MFE | -36.12 |
| Consensus MFE | -27.30 |
| Energy contribution | -27.08 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837169 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16502692 96 - 27905053 AAGACGAUGGCUCCUACAAGCCGCCUUCGCCCCCCGAAGAUGGCUCGUGGCAGCCACAGCCUGGUCUGGAGGGAGAGUACUCGGACCCGGAACUAA .....(.(((((.((((.(((((.(((((.....))))).))))).)))).))))))..((.((((((.((........)))))))).))...... ( -39.50) >DroVir_CAF1 64266 89 - 1 AGGAUGAUGGCAGCUAUAAGCCGCCAAUUGACGCAGAUGAUGGUUUGUGGAAACCACAGCCGGGCCUGGAGGGCGAAUAUGUGGAGUCC------- .((((..((((.((.....)).)))).((..(((((((....)))))))..))(((((.....(((.....))).....))))).))))------- ( -29.50) >DroSec_CAF1 58960 96 - 1 AAGACGAUGGCUCCUACAAGCCGCCUUCGCCACCAGAAGAUGGCUCGUGGCAGCCACAGCCUGGUCUGGAGGGAGAGUACUCGGACCCGGAAUUAA .....(.(((((.((((.(((((.((((.......)))).))))).)))).))))))..((.((((((.((........)))))))).))...... ( -38.30) >DroSim_CAF1 62986 96 - 1 AAGACGAUGGCUCCUACAAGCCGCCUUCGCCACCAGAAGAUGGCUCGUGGCAGCCACAGCCUGGUCUGGAGGGAGAGUACUCGGACCCGGAAUUAA .....(.(((((.((((.(((((.((((.......)))).))))).)))).))))))..((.((((((.((........)))))))).))...... ( -38.30) >DroEre_CAF1 60770 96 - 1 AAGACGAUGGCUCCUACAAGCCGCCUUUGCCCCCCGCAGAUGGCUCGUGGCAGCCGCAGCCUGGUCUGGAGGGAGAGUACUCCGACCCUGAAUUAA ........((((.((((.(((((..(((((.....)))))))))).)))).)))).(((...((((.((((........)))))))))))...... ( -36.10) >DroAna_CAF1 56438 96 - 1 ACGAUGAUGGCUCGUACAAGCCACCCUCGAGUGCGGAGGAUGGCUUAUGGAAGCCCCAGCCGGGCUUGGAGGGAGAGUACGUGGACCCCGAAUUGA .((((...(((((((((...(..((..(((((.(((..(..(((((....))))).)..))).)))))..))..).))))).)).))....)))). ( -35.00) >consensus AAGACGAUGGCUCCUACAAGCCGCCUUCGCCCCCAGAAGAUGGCUCGUGGCAGCCACAGCCUGGUCUGGAGGGAGAGUACUCGGACCCGGAAUUAA .......(((((.((((.(((((.((((.......)))).))))).)))).)))))....((((..(.(((........))).)..))))...... (-27.30 = -27.08 + -0.22)


| Location | 16,502,748 – 16,502,862 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.30 |
| Mean single sequence MFE | -44.47 |
| Consensus MFE | -18.03 |
| Energy contribution | -20.59 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.800249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16502748 114 + 27905053 UCUUCGGGGGGCGAAGGCGGCUUGUAGGAGCCAUCGUCUUCCGGCGGAGGAGGAGGUGGCUGGACUAGCAAGACAACGGGUG------GAGACAGCAGAUAGGUUCCGACUGCAGGAAGA ((((((..(.((....))..(((((((.(((((((.((((((......)))))))))))))...)).))))).)..)...((------....))((((...(....)..))))..))))) ( -42.00) >DroGri_CAF1 75514 111 + 1 UCAUCCUCGCUGAUUGACGGCUUAUAGCUGCCAUCGUCCUCGGGCGCA---------GGCUGCACCAGCAGCAAAACUGGCGUAGCUGGCGCCAACAAAUAAGUGCCAGCCACUGGAAGA ((.(((..((((...((.(((........))).)).......(((((.---------((((((.((((........)))).)))))).))))).............))))....))).)) ( -42.30) >DroSec_CAF1 59016 114 + 1 UCUUCUGGUGGCGAAGGCGGCUUGUAGGAGCCAUCGUCUUCCGGUGGAGGAGGAGGUGGCUGGACUAGCAAGACAACGGGUG------GAGACAGUAGAUAAGUUCCGACUGCUGGAAGG .((((..(..(((....((.(((((((.(((((((.((((((......)))))))))))))...)).)))))....))(.((------....)).)..........)).)..)..)))). ( -44.20) >DroSim_CAF1 63042 114 + 1 UCUUCUGGUGGCGAAGGCGGCUUGUAGGAGCCAUCGUCUUCCGGCGGAGGAGGAGGUGGCUGGACUAGCAAGACAACGGGUG------GAGACAGUAGAUAAGUUCCGACUGCUGGAAGG .((((..(..(((....((.(((((((.(((((((.((((((......)))))))))))))...)).)))))....))(.((------....)).)..........)).)..)..)))). ( -44.20) >DroEre_CAF1 60826 114 + 1 UCUGCGGGGGGCAAAGGCGGCUUGUAGGAGCCAUCGUCUUCCGGCGGAGGAGGAGGUGGUUGGACUAGCAGAACAACGGGUG------GAGACAGCAGAUAGGUUCCGACUGCAGGAAGG ((((((((((((....((.(((.((...(((((((.((((((......)))))))))))))..)).)))...........((------....))))......)))))..))))))).... ( -42.70) >DroAna_CAF1 56494 120 + 1 UCCUCCGCACUCGAGGGUGGCUUGUACGAGCCAUCAUCGUCUGGAGGAGGCGGUGGUGUCUGGACGAGUAAAAGCACUGGUGCUGGCUGGGAAAUCAAGAAGGUUCCAGCCCCCGGCAAG (((((((....(((.((((((((....)))))))).)))..)))))))(.(((.((.(.((((((.(((...((((....)))).))).)...(((.....)))))))))))))).)... ( -51.40) >consensus UCUUCCGGGGGCGAAGGCGGCUUGUAGGAGCCAUCGUCUUCCGGCGGAGGAGGAGGUGGCUGGACUAGCAAGACAACGGGUG______GAGACAGCAGAUAAGUUCCGACUGCUGGAAGG (((((.......))))).(((((....)))))....(((((((((((....((((.(.((((..............................)))).....).))))..))))))))))) (-18.03 = -20.59 + 2.56)

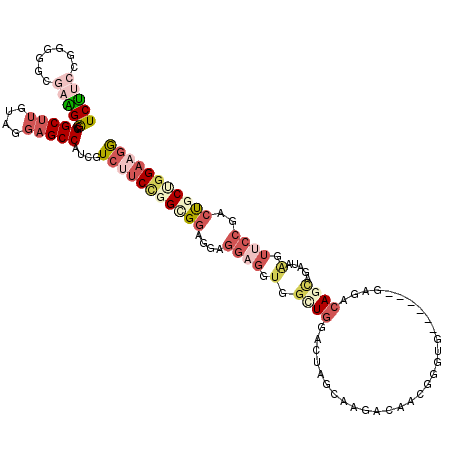
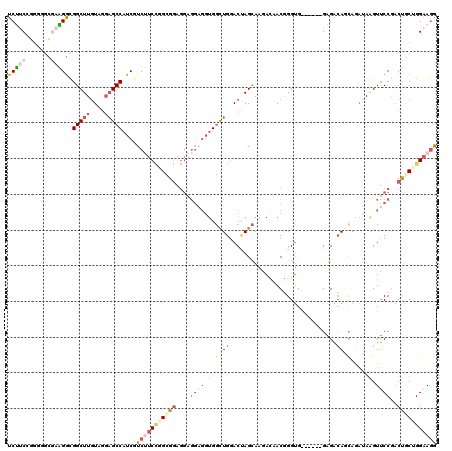
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:18:35 2006