
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,485,086 – 16,485,222 |
| Length | 136 |
| Max. P | 0.998835 |

| Location | 16,485,086 – 16,485,200 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 67.11 |
| Mean single sequence MFE | -24.18 |
| Consensus MFE | -6.50 |
| Energy contribution | -8.18 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.27 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.935474 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16485086 114 + 27905053 AUUACUUAUUCAAUUAUAUCACUUUUCCGCCUUCACCACACUUCACAAUAGGUCGAAUGCAGGUGGAUAAAGGUUCCACCUGGAUUGCUCUUUCGU-UGGAUAAGGCGGUCAAAA ..........................(((((((..((((((((......))))..(((.((((((((.......)))))))).)))........).-)))..)))))))...... ( -30.60) >DroSec_CAF1 41061 95 + 1 ACUACAAAUUCAAUUAUAGCACUUUUCCGCCAUCUCAACACUUCACAAUAAGUCGAAAGCAGGUGGAAAAAGGUUCCACU-------CUCUUUC-------------GGUCAAAA ............................(((........((((......)))).(((((..(((((((.....)))))))-------..)))))-------------)))..... ( -17.30) >DroSim_CAF1 42352 102 + 1 ACUACUUAUUCAAUUAUAGCACUUUGCCGCCUUCUCCACACUUCACAAUAAGUCGAAAGCAGGUGGAAAAAGGUUCCACCUGGAUUACUCUUUC-------------GGUCAAAA ......................((((((...........((((......)))).((((((((((((((.....))))))))).......)))))-------------)).)))). ( -21.11) >DroEre_CAF1 43065 105 + 1 -UUACUUAUCCCAUAGAACCACUU---------CUCCACACUUUACGAUGAGUCGAGUGCAGGUGGAACAAGGAUCCACCUGGAUUGAGUUUUCGUUUGGAUAAGGCAGUCAAAA -...(((((((....(((..((((---------.(((.(((((.((.....)).))))).(((((((.......))))))))))..)))).)))....))))))).......... ( -32.00) >DroYak_CAF1 44017 94 + 1 --UACUUAUUCCAUUGACCCACUUUUCCUCCUUCCCCACACUUCACG-------------------AACAAGGAUCCACCUGGAUUUCCUUUUCGUUUGGAUAAGGCAGUCGAAA --...........(((((...(((.(((................(((-------------------((.((((((((....)))..))))))))))..))).)))...))))).. ( -19.87) >consensus ACUACUUAUUCAAUUAUACCACUUUUCCGCCUUCUCCACACUUCACAAUAAGUCGAAAGCAGGUGGAAAAAGGUUCCACCUGGAUUGCUCUUUCGU_UGGAUAAGGCGGUCAAAA ...........................................................((((((((.......))))))))................................. ( -6.50 = -8.18 + 1.68)



| Location | 16,485,086 – 16,485,200 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 67.11 |
| Mean single sequence MFE | -28.74 |
| Consensus MFE | -5.44 |
| Energy contribution | -7.44 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.19 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.564422 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16485086 114 - 27905053 UUUUGACCGCCUUAUCCA-ACGAAAGAGCAAUCCAGGUGGAACCUUUAUCCACCUGCAUUCGACCUAUUGUGAAGUGUGGUGAAGGCGGAAAAGUGAUAUAAUUGAAUAAGUAAU ......(((((((.....-.((((.((....))((((((((.......))))))))..))))(((((((....)))).))).))))))).......................... ( -32.50) >DroSec_CAF1 41061 95 - 1 UUUUGACC-------------GAAAGAG-------AGUGGAACCUUUUUCCACCUGCUUUCGACUUAUUGUGAAGUGUUGAGAUGGCGGAAAAGUGCUAUAAUUGAAUUUGUAGU (((..(((-------------(((((((-------.((((((.....)))))))).))))))((((......))))))..)))............(((((((......))))))) ( -24.70) >DroSim_CAF1 42352 102 - 1 UUUUGACC-------------GAAAGAGUAAUCCAGGUGGAACCUUUUUCCACCUGCUUUCGACUUAUUGUGAAGUGUGGAGAAGGCGGCAAAGUGCUAUAAUUGAAUAAGUAGU (((((.((-------------....((....)).((((((((.....))))))))((((((..(.((((....)))).)..))))))))))))).(((((..........))))) ( -27.80) >DroEre_CAF1 43065 105 - 1 UUUUGACUGCCUUAUCCAAACGAAAACUCAAUCCAGGUGGAUCCUUGUUCCACCUGCACUCGACUCAUCGUAAAGUGUGGAG---------AAGUGGUUCUAUGGGAUAAGUAA- ..........(((((((....(((.(((...((((((((((.......)))))))(((((..((.....))..)))))))).---------.)))..)))....)))))))...- ( -33.80) >DroYak_CAF1 44017 94 - 1 UUUCGACUGCCUUAUCCAAACGAAAAGGAAAUCCAGGUGGAUCCUUGUU-------------------CGUGAAGUGUGGGGAAGGAGGAAAAGUGGGUCAAUGGAAUAAGUA-- ..((.(((.((((.((((.((((((((((..(((....)))))))).))-------------------)))......))))....))))...))).))...............-- ( -24.90) >consensus UUUUGACCGCCUUAUCCA_ACGAAAGAGCAAUCCAGGUGGAACCUUUUUCCACCUGCAUUCGACUUAUUGUGAAGUGUGGAGAAGGCGGAAAAGUGCUAUAAUUGAAUAAGUAAU .................................((((((((.......))))))))........................................................... ( -5.44 = -7.44 + 2.00)



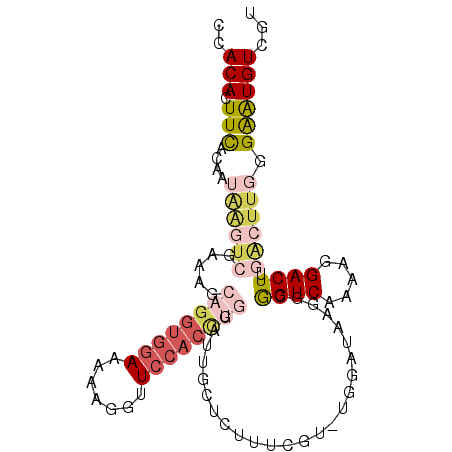
| Location | 16,485,121 – 16,485,222 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 67.86 |
| Mean single sequence MFE | -30.48 |
| Consensus MFE | -12.14 |
| Energy contribution | -14.46 |
| Covariance contribution | 2.32 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.40 |
| SVM decision value | 3.25 |
| SVM RNA-class probability | 0.998835 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16485121 101 + 27905053 CCACACUUCACAAUAGGUCGAAUGCAGGUGGAUAAAGGUUCCACCUGGAUUGCUCUUUCGU-UGGAUAAGGCGGUCAAAAGGACUUGACUUGGGAGUGUCAU ..(((((((.(((((((((.....((((((((.......))))))))((((((.(((((..-..)).))))))))).....))))))..))))))))))... ( -38.40) >DroSec_CAF1 41096 82 + 1 CAACACUUCACAAUAAGUCGAAAGCAGGUGGAAAAAGGUUCCACU-------CUCUUUC-------------GGUCAAAAGGACUUGACUUGGGAAUGUCGU ..(((.(((....(((((((((((..(((((((.....)))))))-------..)))))-------------((((.....)))).)))))).))))))... ( -25.60) >DroSim_CAF1 42387 89 + 1 CCACACUUCACAAUAAGUCGAAAGCAGGUGGAAAAAGGUUCCACCUGGAUUACUCUUUC-------------GGUCAAAAGGACUUGACUUGGGAAUGUCGU ..(((.(((....((((((((((((((((((((.....))))))))).......)))))-------------((((.....)))).)))))).))))))... ( -27.91) >DroEre_CAF1 43090 102 + 1 CCACACUUUACGAUGAGUCGAGUGCAGGUGGAACAAGGAUCCACCUGGAUUGAGUUUUCGUUUGGAUAAGGCAGUCAAAAGGACUGUGGUUUGGGAUGUUUG (((..((..((((.((.((.(((.((((((((.......)))))))).))))).)).))))..)).....((((((.....))))))....)))........ ( -35.20) >DroYak_CAF1 44050 81 + 1 CCACACUUCACG-------------------AACAAGGAUCCACCUGGAUUUCCUUUUCGUUUGGAUAAGGCAGUCGAAAGGACUUAAGCUAGGAGUGUC-- ..((((((((((-------------------((.((((((((....)))..))))))))))........(((((((.....))))...))).))))))).-- ( -25.30) >consensus CCACACUUCACAAUAAGUCGAAAGCAGGUGGAAAAAGGUUCCACCUGGAUUGCUCUUUCGU_UGGAUAAGGCGGUCAAAAGGACUUGACUUGGGAAUGUCGU ..(((.(((....((((((.....((((((((.......)))))))).........................((((.....)))).)))))).))))))... (-12.14 = -14.46 + 2.32)


| Location | 16,485,121 – 16,485,222 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 67.86 |
| Mean single sequence MFE | -25.34 |
| Consensus MFE | -9.24 |
| Energy contribution | -11.20 |
| Covariance contribution | 1.96 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.36 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16485121 101 - 27905053 AUGACACUCCCAAGUCAAGUCCUUUUGACCGCCUUAUCCA-ACGAAAGAGCAAUCCAGGUGGAACCUUUAUCCACCUGCAUUCGACCUAUUGUGAAGUGUGG ...(((((.((((((((((....))))))...........-.((((.((....))((((((((.......))))))))..)))).....))).).))))).. ( -27.60) >DroSec_CAF1 41096 82 - 1 ACGACAUUCCCAAGUCAAGUCCUUUUGACC-------------GAAAGAG-------AGUGGAACCUUUUUCCACCUGCUUUCGACUUAUUGUGAAGUGUUG .(((((((.((((((((((....))))))(-------------(((((((-------.((((((.....)))))))).)))))).....))).).))))))) ( -27.40) >DroSim_CAF1 42387 89 - 1 ACGACAUUCCCAAGUCAAGUCCUUUUGACC-------------GAAAGAGUAAUCCAGGUGGAACCUUUUUCCACCUGCUUUCGACUUAUUGUGAAGUGUGG .(.(((((.((((((((((....))))))(-------------(((((.......(((((((((.....))))))))))))))).....))).).))))).) ( -28.41) >DroEre_CAF1 43090 102 - 1 CAAACAUCCCAAACCACAGUCCUUUUGACUGCCUUAUCCAAACGAAAACUCAAUCCAGGUGGAUCCUUGUUCCACCUGCACUCGACUCAUCGUAAAGUGUGG ................(((((.....)))))......(((...............((((((((.......))))))))((((..((.....))..))))))) ( -23.60) >DroYak_CAF1 44050 81 - 1 --GACACUCCUAGCUUAAGUCCUUUCGACUGCCUUAUCCAAACGAAAAGGAAAUCCAGGUGGAUCCUUGUU-------------------CGUGAAGUGUGG --.(((((.(..((...((((.....)))))).........((((((((((..(((....)))))))).))-------------------)))).))))).. ( -19.70) >consensus ACGACAUUCCCAAGUCAAGUCCUUUUGACCGCCUUAUCCA_ACGAAAGAGCAAUCCAGGUGGAACCUUUUUCCACCUGCAUUCGACUUAUUGUGAAGUGUGG ...(((((..........(((.....)))..........................((((((((.......)))))))).................))))).. ( -9.24 = -11.20 + 1.96)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:18:22 2006