
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,450,208 – 16,450,310 |
| Length | 102 |
| Max. P | 0.995476 |

| Location | 16,450,208 – 16,450,310 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 76.05 |
| Mean single sequence MFE | -34.84 |
| Consensus MFE | -18.00 |
| Energy contribution | -17.83 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.40 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.52 |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.995476 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
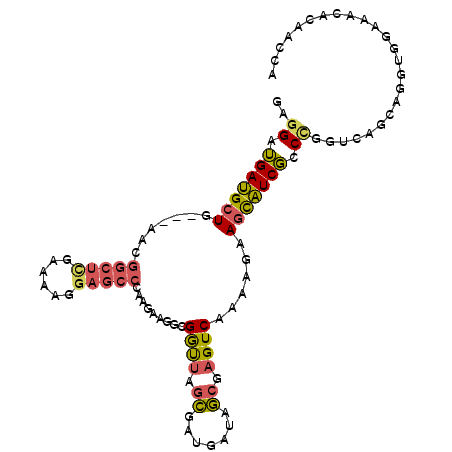
>3R_DroMel_CAF1 16450208 102 + 27905053 GAGGAUGAUGCUG---AACGGCUCGAAAAGGAGCCGAAGAAGGCGGUUAGCGAUGAUAGCGAGUCAAAAGAAGCAUCGCCCGGUCAGCAAGUGGAACCACAACCA ..((....(((((---(.((((((......)))))).....((((((..((.......))(..((....))..)))))))...)))))).(((....)))..)). ( -36.40) >DroSec_CAF1 6287 102 + 1 GAGGAUGAUGCUG---AACGGCUCGAAAUGGAGCCGAAGAAGGCGGCUAGCGAUGAUAGCGAGUCAAAAGAAGCAUCGCCUGGUCAGCAGGUGGAAACACAACCA ..((....(((((---(.((((((......))))))....(((((((((.......))))(..((....))..)..)))))..)))))).(((....)))..)). ( -41.00) >DroSim_CAF1 6334 102 + 1 GAGGAUGAUGCUG---AACGGCUCGAAAUGGAGCCUAAGAAGGCGACUAGCGAUGAUAGCGAGUCAAAAGAAGCAUCGCCUGGUCAGCAGGUGGAAACACAACCA ..((....(((((---(.((((((......))))).....((((((((.((.......)).))))....((....))))))).)))))).(((....)))..)). ( -38.60) >DroEre_CAF1 6209 102 + 1 GACGAUGAUGCUG---AACGGCUUGAAAAGGAGCCCAAGAAAGCGGUUAGCGAUGACAGCGAGUCAAAAACAGCAUCGCCCGCUCAGCACGGCGAUACACAACCA ..((.((.(((((---(..(((((......))))).......((((...((((((..................)))))))))))))))))).))........... ( -29.57) >DroYak_CAF1 6931 102 + 1 CAGGAUGAUGCUG---AACGGCUUGAAAAUGAGCCAAAGACGGCGGUUAGCGAUGACAGCGAGUCAAAAACAGCAUCGCCCGCUCAGCAGGUUGAUACAAAACCA ..((.((((((((---...(((((......)))))...(((.((.((((....)))).))..))).....)))))))).))........((((.......)))). ( -35.60) >DroAna_CAF1 5272 99 + 1 GAGGAGGAAGAUGGCACAUUGCCGGUAAAGGACGUCCAACCGGAGGUCAAGGAGAAGGCCGAGUCAGAAGCAGUUUCUCCCA------AAGCAGAGCCGGAGGCA (.((((((((.((((......(((((...((....)).))))).((((........))))..))))....)..)))))))).------.......(((...))). ( -27.90) >consensus GAGGAUGAUGCUG___AACGGCUCGAAAAGGAGCCCAAGAAGGCGGUUAGCGAUGAUAGCGAGUCAAAAGAAGCAUCGCCCGGUCAGCAGGUGGAAACACAACCA ..((.(((((((.......(((((......))))).........((((.((.......)).))))......))))))).))........................ (-18.00 = -17.83 + -0.16)

| Location | 16,450,208 – 16,450,310 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 76.05 |
| Mean single sequence MFE | -29.83 |
| Consensus MFE | -12.82 |
| Energy contribution | -13.94 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563476 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
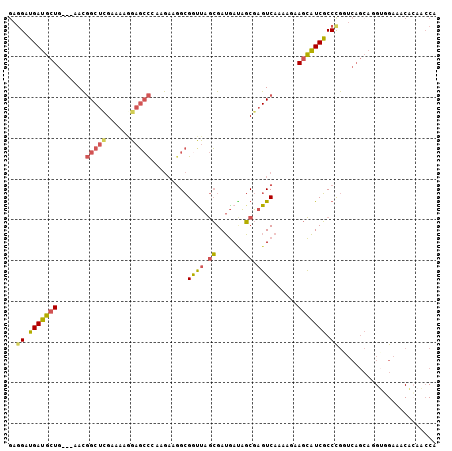
>3R_DroMel_CAF1 16450208 102 - 27905053 UGGUUGUGGUUCCACUUGCUGACCGGGCGAUGCUUCUUUUGACUCGCUAUCAUCGCUAACCGCCUUCUUCGGCUCCUUUUCGAGCCGUU---CAGCAUCAUCCUC .((..(((....))).((((((.(((((((((..................))))))...))).......((((((......)))))).)---)))))....)).. ( -29.17) >DroSec_CAF1 6287 102 - 1 UGGUUGUGUUUCCACCUGCUGACCAGGCGAUGCUUCUUUUGACUCGCUAUCAUCGCUAGCCGCCUUCUUCGGCUCCAUUUCGAGCCGUU---CAGCAUCAUCCUC .(((.(.....).)))((((((..(((((..(((.....(((.......))).....))))))))....((((((......)))))).)---)))))........ ( -29.20) >DroSim_CAF1 6334 102 - 1 UGGUUGUGUUUCCACCUGCUGACCAGGCGAUGCUUCUUUUGACUCGCUAUCAUCGCUAGUCGCCUUCUUAGGCUCCAUUUCGAGCCGUU---CAGCAUCAUCCUC .(((.(.....).)))((((((.(((((((....))....((((.((.......)).)))))))).....(((((......)))))).)---)))))........ ( -30.40) >DroEre_CAF1 6209 102 - 1 UGGUUGUGUAUCGCCGUGCUGAGCGGGCGAUGCUGUUUUUGACUCGCUGUCAUCGCUAACCGCUUUCUUGGGCUCCUUUUCAAGCCGUU---CAGCAUCAUCGUC .(((........)))((((((((((((((((((.((.........)).).)))))).....((((....))))...........)))))---))))))....... ( -31.80) >DroYak_CAF1 6931 102 - 1 UGGUUUUGUAUCAACCUGCUGAGCGGGCGAUGCUGUUUUUGACUCGCUGUCAUCGCUAACCGCCGUCUUUGGCUCAUUUUCAAGCCGUU---CAGCAUCAUCCUG .((((.......))))(((((((((((((((((.((.........)).).)))))).....((((....))))...........)))))---)))))........ ( -32.90) >DroAna_CAF1 5272 99 - 1 UGCCUCCGGCUCUGCUU------UGGGAGAAACUGCUUCUGACUCGGCCUUCUCCUUGACCUCCGGUUGGACGUCCUUUACCGGCAAUGUGCCAUCUUCCUCCUC .(((...))).......------.((((((....((.((..((.(((...((.....))...)))))..)).))........(((.....))).))))))..... ( -25.50) >consensus UGGUUGUGUUUCCACCUGCUGACCGGGCGAUGCUGCUUUUGACUCGCUAUCAUCGCUAACCGCCUUCUUCGGCUCCUUUUCGAGCCGUU___CAGCAUCAUCCUC ................(((((....(((((((..................))))))).............((((........))))......)))))........ (-12.82 = -13.94 + 1.11)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:18:11 2006