
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,447,535 – 16,447,720 |
| Length | 185 |
| Max. P | 0.899022 |

| Location | 16,447,535 – 16,447,648 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.10 |
| Mean single sequence MFE | -31.06 |
| Consensus MFE | -17.74 |
| Energy contribution | -18.86 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711336 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16447535 113 + 27905053 GC------AGCGACCUGUGUGUAAGCUUACAUUGAGAUAACAUUAAUGUGAACGUGCAGCGAAUGCAGC-ACUAUAACAUUGGCCAGUGGAAACAUGAUAUUUGCUAGCAUGCUGACUGG .(------(((..((((..(((....(((((((((.......))))))))))))..))).).(((((((-((((......)))...(((....)))......)))).))))))))..... ( -32.00) >DroSec_CAF1 3563 120 + 1 GCAAUAACAGCAACCUGGGUGUAAGUUUACAUUAAGAUAACAUUAAUGUGAACGUGCAGCGAAUGCAGCUACUAUAACAUUGGCCAGUGGAAACAUGACAUUUGCUAGCAUGCUGACUGG .......((((.............(((((((((((.......)))))))))))(((((((((((((.((((.........))))..(((....)))).)))))))).))))))))..... ( -37.10) >DroSim_CAF1 3611 120 + 1 GCAAUAACAGCAACCUGGGUGUAAGCUUACAUUAAGAUAACAUUAAUGUGAACGUGCAGCGAAUGCAGCCACUAUAGCAUUGGCCAGUGGAAACAUGACAUUUGCUAGCAUGCUGACUGG .......((((.............(.(((((((((.......))))))))).)((((((((((((((.(((((...((....)).))))).....)).)))))))).))))))))..... ( -35.80) >DroEre_CAF1 3592 97 + 1 GCAU-----------------------AACAUUAACAUAACAUUAAUGUGAACGUUCGGUAAAUAAAACCACUAAAAUCUUGGCCAGUGGAAACAUGAUAUUUGCUAGCAUGCUGACUGG ((((-----------------------.(((((((.......)))))))....(((.((((((((...(((.........)))...(((....)))..)))))))))))))))....... ( -18.90) >DroYak_CAF1 4285 119 + 1 GCAUCAACAGCGACCUGUGUGUUAGCUUACAUUAACAUAACAUAAAUGUAAACGUGUGGUUAAUGCAACCAGUUA-ACAUUGGCCAGUGGAAAGUGGAUAUUUGUUAGCAUGCUGACUGG .......(((((.....((((((((((.((((..((((.......))))....))))))))))))))....((((-(((....(((.(....).))).....))))))).)))))..... ( -31.50) >consensus GCAA_AACAGCAACCUGGGUGUAAGCUUACAUUAAGAUAACAUUAAUGUGAACGUGCAGCGAAUGCAGCCACUAUAACAUUGGCCAGUGGAAACAUGAUAUUUGCUAGCAUGCUGACUGG .......((((...............(((((((((.......)))))))))..((((((((((((((.(((((............))))).....)).)))))))).))))))))..... (-17.74 = -18.86 + 1.12)

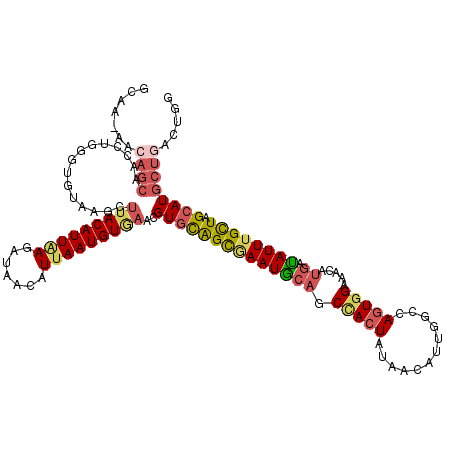

| Location | 16,447,569 – 16,447,686 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 89.49 |
| Mean single sequence MFE | -30.48 |
| Consensus MFE | -24.50 |
| Energy contribution | -24.38 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.520549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16447569 117 - 27905053 UGUGUGACCGUCAACUAAGAACGUCAAAAUCCACGGCGCCAGUCAGCAUGCUAGCAAAUAUCAUGUUUCCACUGGCCAAUGUUAUAGU-GCUGCAUUCGCUGCACGUUCACAUUAAUG .((((((.(((..((((..((((((.........)))((((((.((((((.((.....)).))))))...))))))....))).))))-((.((....)).))))).))))))..... ( -31.30) >DroSec_CAF1 3603 118 - 1 UGUGUGACCGUCAACUAAGAACGUCAAAAUCCACGGCGCCAGUCAGCAUGCUAGCAAAUGUCAUGUUUCCACUGGCCAAUGUUAUAGUAGCUGCAUUCGCUGCACGUUCACAUUAAUG .((((((.(((..((((..((((((.........)))((((((.(((((((........).))))))...))))))....))).)))).((.((....)).))))).))))))..... ( -31.80) >DroSim_CAF1 3651 118 - 1 UGUGUGACCGUCAACUAAGAACGUCAAAAUCCACGGCGCCAGUCAGCAUGCUAGCAAAUGUCAUGUUUCCACUGGCCAAUGCUAUAGUGGCUGCAUUCGCUGCACGUUCACAUUAAUG .((((((.(((..((((.....(((.........)))((((((.(((((((........).))))))...))))))........)))).((.((....)).))))).))))))..... ( -30.50) >DroEre_CAF1 3609 118 - 1 UGUGUGACCGUCAACUAAGAACGUCAAAAUCGACAGCGCCAGUCAGCAUGCUAGCAAAUAUCAUGUUUCCACUGGCCAAGAUUUUAGUGGUUUUAUUUACCGAACGUUCACAUUAAUG .((((((.((((.(((((((.((((......))).(.((((((.((((((.((.....)).))))))...)))))))..).)))))))(((.......)))).))).))))))..... ( -33.90) >DroYak_CAF1 4325 117 - 1 UGUGUGACCGUCAACUAAGAACGUAGAAAUCCACGGCGCCAGUCAGCAUGCUAACAAAUAUCCACUUUCCACUGGCCAAUGU-UAACUGGUUGCAUUAACCACACGUUUACAUUUAUG .(((((...............(((.(....).)))((((((((.(((((((((...................))))..))))-).))))).)))......)))))............. ( -24.91) >consensus UGUGUGACCGUCAACUAAGAACGUCAAAAUCCACGGCGCCAGUCAGCAUGCUAGCAAAUAUCAUGUUUCCACUGGCCAAUGUUAUAGUGGCUGCAUUCGCUGCACGUUCACAUUAAUG .((((((.(((........................(.((((((.((((((.((.....)).))))))...))))))).........(((((.......)))))))).))))))..... (-24.50 = -24.38 + -0.12)



| Location | 16,447,608 – 16,447,720 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 96.25 |
| Mean single sequence MFE | -28.44 |
| Consensus MFE | -24.84 |
| Energy contribution | -25.28 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16447608 112 - 27905053 AAAAAUUAUCAACGAAAAACGCAGCUCGGUGCGCUGUGUGACCGUCAACUAAGAACGUCAAAAUCCACGGCGCCAGUCAGCAUGCUAGCAAAUAUCAUGUUUCCACUGGCCA ....................(((......)))((((((((((..((......))..)))).....))))))((((((.((((((.((.....)).))))))...)))))).. ( -28.40) >DroSec_CAF1 3643 112 - 1 AAAAAUUAUCAACGAAAAACGCAGCUCGGUGCGCUGUGUGACCGUCAACUAAGAACGUCAAAAUCCACGGCGCCAGUCAGCAUGCUAGCAAAUGUCAUGUUUCCACUGGCCA ....................(((......)))((((((((((..((......))..)))).....))))))((((((.(((((((........).))))))...)))))).. ( -28.70) >DroSim_CAF1 3691 112 - 1 AAAAAUUAUCAACGAAAAACGCAGCUCGGUGCGCUGUGUGACCGUCAACUAAGAACGUCAAAAUCCACGGCGCCAGUCAGCAUGCUAGCAAAUGUCAUGUUUCCACUGGCCA ....................(((......)))((((((((((..((......))..)))).....))))))((((((.(((((((........).))))))...)))))).. ( -28.70) >DroEre_CAF1 3649 112 - 1 AAAAAUUAUCAACGAAAAACGCAGCUCGGUGCGCUGUGUGACCGUCAACUAAGAACGUCAAAAUCGACAGCGCCAGUCAGCAUGCUAGCAAAUAUCAUGUUUCCACUGGCCA ............(((...(((((((.......)))))))(((..((......))..)))....)))...(.((((((.((((((.((.....)).))))))...))))))). ( -29.40) >DroYak_CAF1 4364 112 - 1 AAAAAUUAUCAACGAAAAACGCAGCUCUGUGCGCUGUGUGACCGUCAACUAAGAACGUAGAAAUCCACGGCGCCAGUCAGCAUGCUAACAAAUAUCCACUUUCCACUGGCCA ....................(((((((((.((((((((.((...((.((.......)).))..)))))))))))))..))).)))..........(((........)))... ( -27.00) >consensus AAAAAUUAUCAACGAAAAACGCAGCUCGGUGCGCUGUGUGACCGUCAACUAAGAACGUCAAAAUCCACGGCGCCAGUCAGCAUGCUAGCAAAUAUCAUGUUUCCACUGGCCA ....................(((......)))((((((((((..((......))..)))).....))))))((((((.((((((.((.....)).))))))...)))))).. (-24.84 = -25.28 + 0.44)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:18:07 2006