
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,440,490 – 16,440,648 |
| Length | 158 |
| Max. P | 0.764858 |

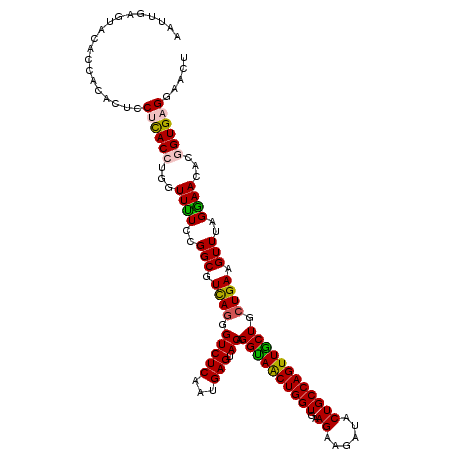
| Location | 16,440,490 – 16,440,609 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 81.83 |
| Mean single sequence MFE | -38.58 |
| Consensus MFE | -29.81 |
| Energy contribution | -29.32 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16440490 119 + 27905053 AAUUGAGUACAUCACACUCCUCACCUGGUUUUCCGGCGUCAGGGUCUCGAUGAGUACAGGUAACUGGUGAAGCAAAUACUGCCAGUUACUACUGAAGUUUAGAAACACGGUGAGAAACU ....((((.......))))((((((..(((((..(((.((((.(((((...))).)).((((((((((............)))))))))).)))).)))..)))))..))))))..... ( -37.30) >DroPse_CAF1 37837 112 + 1 AAUGGAUCAAAU-------CCCACCUGGUUCUCUGGCGUCAGAGUCUCGAUGAGCACGGGCAGCUGGUGGAGAAGGUGCUGCCAGUUGCUGCUGAAGUUGAGGAAGAGCGUGAGGUAGU ............-------(((((((..(((((.(((.((((.(((((...))).)).(((((((((..(........)..))))))))).)))).))))))))..)).))).)).... ( -39.00) >DroSec_CAF1 36417 119 + 1 AAUUGAGUACACCACACUCCUCACCUGGUUUUCCGGCGUCAGGGUCUCAAUGAGUACAGGUAGCUGGUGAAGCAGAUACUGCCAGUUGCUGCUGAAGUUUAGGAACACGGUGAGGAACU .................((((((((..((.((((.((.((((.(((((...))).)).((((((((((............)))))))))).)))).))...)))).))))))))))... ( -39.60) >DroSim_CAF1 36912 119 + 1 AAUUGAGUACACCACACUCCUCACCUGGUUUUCCGGCGUCAGGGUCUCAAUGAGUACAGGUAACUGGUGAAGCAAAUACUGCCAGUUGCUGCUGAAGUUUAGGAACACGGUGAGGAACU .................((((((((..((.((((.((.((((.(((((...))).)).((((((((((............)))))))))).)))).))...)))).))))))))))... ( -39.60) >DroEre_CAF1 41284 119 + 1 CAUUGAAUAUACCACUCUCCUUACCUGGUUUUCCGGCGUUAAUGUCUCUAUGAGUACAGGUAACUGGUGAAGAAGAUACUGCCAGUUACUGCUGAAGUUUAGGAAAACGGUGAGGAACU .................((((((((..(((((((.((.....((((((...))).)))((((((((((............))))))))))......))...)))))))))))))))... ( -36.80) >DroPer_CAF1 37710 112 + 1 AAUGGAUCGAAU-------CCCACCUGGUUCUCUGGCGUCAGGGUCUCAAUGAGCACGGGCAGCUGGUGGAGAAGGUGCUGCCAGUUGCUGCUGAAGUUGAGGAAGAGCGUGAGGUAGU ............-------(((((((..(((((.(((.(((((.((.....)).)...(((((((((..(........)..))))))))).)))).))))))))..)).))).)).... ( -39.20) >consensus AAUUGAGUACACCACACUCCUCACCUGGUUUUCCGGCGUCAGGGUCUCAAUGAGUACAGGUAACUGGUGAAGAAGAUACUGCCAGUUGCUGCUGAAGUUUAGGAACACGGUGAGGAACU ...................((((((...((((..(((.((((.(((((...))).)).((((((((((..((......)))))))))))).)))).)))..))))...))))))..... (-29.81 = -29.32 + -0.50)

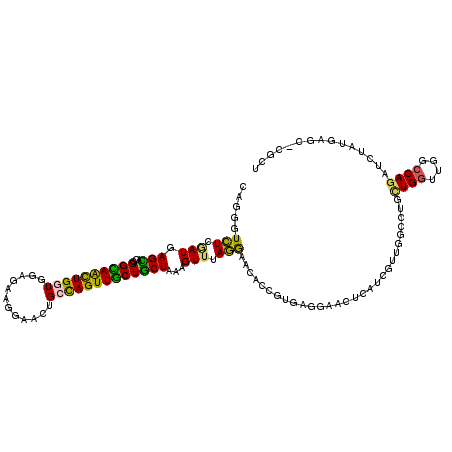
| Location | 16,440,529 – 16,440,648 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.42 |
| Mean single sequence MFE | -41.37 |
| Consensus MFE | -19.02 |
| Energy contribution | -17.72 |
| Covariance contribution | -1.30 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.662955 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16440529 119 + 27905053 CAGGGUCUCGAUGAGUACAGGUAACUGGUGAAGCAAAUACUGCCAGUUACUACUGAAGUUUAGAAACACGGUGAGAAACUCGGCGUUGUGCCGUUGGUUGGCCAGGUCAAUGAGA-CGCU .((.((((((.(((.....((((((((((............)))))))))).(((..........(((((.((((...)))).)).)))((((.....))))))).))).)))))-).)) ( -40.00) >DroPse_CAF1 37869 119 + 1 CAGAGUCUCGAUGAGCACGGGCAGCUGGUGGAGAAGGUGCUGCCAGUUGCUGCUGAAGUUGAGGAAGAGCGUGAGGUAGUUAUCGUUGGCCUGCUGGCCGGCCAGAUCUAUUAUC-CGCU .....(((((((...((..(((((((((..(........)..)))))))))..))..)))))))...((((.(((((((.....(((((((....))))))).....))))).))-)))) ( -48.60) >DroGri_CAF1 44634 119 + 1 UAGUGUCUCGAUGAGCACGGGCAACUGAUGCAGCAGGAACUGAUAGUUGCUGCUAAAGUUUAGAUACAGAUUGAUGUAGUUGUCAUUAGCCUGCUGAUUGGCCACAUCUAUAAGC-UGCU ..((((((.(((..((((((....))).)))(((((.(((.....))).)))))...))).)))))).....(((((....((((((((....)))).)))).))))).......-.... ( -36.50) >DroYak_CAF1 40557 119 + 1 UAGGGUCUCAAUGAGUACAGGUAACUGGUGAAAAAAAAACUGCCAGUUACUGCUAAAGUUUAGGAAAACGGUGAGAAACUCGUCGUUGUGCCGCUGGUUGGCCAGAUCAAUGAGU-CGCU .((.(.((((.(((((((.((((((((((............))))))))))...............((((..(((...)))..))))))))..((((....)))).))).)))).-).)) ( -39.20) >DroAna_CAF1 30448 119 + 1 UAGGGUUUCUAUGAGCACUGGUAGUUGAUGGAGCAGGAACUGCCAGUUGCUGCUAAAGUUCAAGUACACUGUAAAGAACUCAUCAUUGUCCUGCUGGGUAACCAGAUCUAUGAGC-CGCU ...(((((.((.((...(((((...(((((.(((((.(((.....))).)))))..(((((...(((...)))..)))))))))).(..((....))..)))))).)))).))))-)... ( -34.10) >DroPer_CAF1 37742 120 + 1 CAGGGUCUCAAUGAGCACGGGCAGCUGGUGGAGAAGGUGCUGCCAGUUGCUGCUGAAGUUGAGGAAGAGCGUGAGGUAGUUAUCGUUGGCCUGCUGGCCGGCCAGAUCUAUUAUCCCGCU .....(((((((...((..(((((((((..(........)..)))))))))..))..)))))))...((((.(.((((((.((((((((((....)))))))..)))..))))))))))) ( -49.80) >consensus CAGGGUCUCGAUGAGCACGGGCAACUGGUGGAGAAGGAACUGCCAGUUGCUGCUAAAGUUUAGGAACACCGUGAGGAACUCAUCGUUGGCCUGCUGGUUGGCCAGAUCUAUGAGC_CGCU .....(((.(((.(((...((((((((((............)))))))))))))...))).))).............................((((....))))............... (-19.02 = -17.72 + -1.30)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:18:03 2006