
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,433,876 – 16,434,030 |
| Length | 154 |
| Max. P | 0.537198 |

| Location | 16,433,876 – 16,433,993 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 81.42 |
| Mean single sequence MFE | -44.02 |
| Consensus MFE | -27.35 |
| Energy contribution | -27.38 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.62 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507486 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16433876 117 + 27905053 AGCCUGCGGGGCCUACUGAACAACCAGGCGGCGCAGAGCCCGAAUGCCCACAUGGCAUUUAAUGGACCGCCCAGCUAUCCGGGUGGGCCGCAAGGAGCACCGGCUCCGGCCCACCAA ....((..(((((..(((......)))(((((......((.(((((((.....)))))))...)).((((((........)))))))))))..(((((....))))))))))..)). ( -55.20) >DroVir_CAF1 28515 117 + 1 AGCCUGCGUGGUCUGUUGAAUAAUCAGGCGGCGCAGAGUCCUAACGCUCAUCUGGCUUUUAAUGGGCCACCCAGCUAUCCAGGCGGGCCGCAGGGCGCACCCUCGCCGGCACACCAG .((((((((.(((((.........))))).)))))((((......))))....(((........((((..((.........))..))))(.((((....)))))))))))....... ( -44.60) >DroGri_CAF1 36020 117 + 1 AGUUUGCGUGGUCUUUUGAAUAAUCAGGCGGCGCAGAGCCCAAAUGCUCAUAUGGCCUUCAAUGGGCCGCCCAGUUAUCCCGGUGGACCACAGGGUGCACCAUCACCGGCCCAUCAA .....(((((((((.(((.(((((..((((((...((((......))))......((......))))))))..)))))..))).)))))))..((((......)))).))....... ( -42.40) >DroWil_CAF1 32217 117 + 1 AGUCUGCGUGGCCUGCUGAACAAUCAGGCGGCGCAGAGUCCCAAUGCCCAUAUGGCCUUUAAUGGACCUCCCAGCUAUCCAGGUGGACCGCAGGGUGCACCAUCUCCUGCACAUCAG ..(((((((.(((((.(.....).))))).)))))))((((....(((...(((((......(((.....))))))))...))))))).((((((.........))))))....... ( -42.20) >DroMoj_CAF1 27090 117 + 1 AGCCUGCGAGGAUUGCUAAACAACCAAGCAGCACAAAGUCCUAAUGCUCAUUUGGCAUUUAAUGGCCCGCCUAGCUAUCCAGGCGGACCGCAGGGCACACCUUCACCGGCCCACCAG .(((.(.((((.(((((.........)))))......(((((((((((.....))))))....((.((((((........)))))).))..)))))....)))).).)))....... ( -37.00) >DroAna_CAF1 24145 117 + 1 AGCCUGCGGGGAUUGUUAAACAAUCAGGCGGCACAGAGCCCCAAUGCCCACAUGGCAUUCAAUGGCCCGCCCAGCUAUCCCGGCGGACCGCAGGGAGCACCGUCACCGGCCCACCAA .(((.((((((((((.....))))).((.(((.....)))))((((((.....))))))......)))))...((..((((.(((...))).)))))).........)))....... ( -42.70) >consensus AGCCUGCGUGGACUGCUGAACAAUCAGGCGGCGCAGAGCCCCAAUGCCCAUAUGGCAUUUAAUGGACCGCCCAGCUAUCCAGGCGGACCGCAGGGAGCACCAUCACCGGCCCACCAA .(((((((..(((((.........)))))..))))..(((((...(((.....)))......(((.(((((..........))))).)))..))).)).........)))....... (-27.35 = -27.38 + 0.03)



| Location | 16,433,916 – 16,434,030 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 78.95 |
| Mean single sequence MFE | -50.32 |
| Consensus MFE | -25.82 |
| Energy contribution | -26.02 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.537198 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16433916 114 - 27905053 CGGGCAUGAAAUGCGGACGCAUCUGCGGGCCCAUCGGUUGGUGGGCCGGAGCCGGUGCUCCUUGCGGCCCACCCGGAUAGCUGGGCGGUCCAUUAAAUGCCAUGUGGGCAUUCG .((((......((((((....)))))).(((((.(.((((((((((((((((....)))))....))))))))..))).).))))).))))....((((((.....)))))).. ( -53.40) >DroVir_CAF1 28555 114 - 1 CGGGCAUAAAAUGCGCCCGCAUCGGGGGUAAUGAUUGCUGGUGUGCCGGCGAGGGUGCGCCCUGCGGCCCGCCUGGAUAGCUGGGUGGCCCAUUAAAAGCCAGAUGAGCGUUAG (((((.........)))))(((((((.(((....(((((((....)))))))...))).)))...((.(((((..(....)..))))).))...........))))........ ( -48.30) >DroGri_CAF1 36060 114 - 1 CUGGCAUAAAAUGCGGACGCAUCUGUGGCAUUUGCUGUUGAUGGGCCGGUGAUGGUGCACCCUGUGGUCCACCGGGAUAACUGGGCGGCCCAUUGAAGGCCAUAUGAGCAUUUG ........((((((.....((((.(..((....))..).))))((((..((((((.((.(((.((.((((....)))).)).)))..))))))))..))))......)))))). ( -45.20) >DroWil_CAF1 32257 114 - 1 UGGACAGAAAAUGUGGUCGCAUCUGAGGUCCCAUUUGCUGAUGUGCAGGAGAUGGUGCACCCUGCGGUCCACCUGGAUAGCUGGGAGGUCCAUUAAAGGCCAUAUGGGCAUUGG ......(...(((((((((((...(..(..((((((.(((.....))).))))))..)..).)))((.((.((..(....)..)).)).))......))))))))...)..... ( -40.00) >DroMoj_CAF1 27130 114 - 1 CGGGCAUGAAAUGUGGGCGCAUCGGUGGCAUCGAUUGCUGGUGGGCCGGUGAAGGUGUGCCCUGCGGUCCGCCUGGAUAGCUAGGCGGGCCAUUAAAUGCCAAAUGAGCAUUAG ..(((((..(((((((((((((((((...)))..(..((((....))))..).))))))))).))(((((((((((....))))))))))))))..)))))............. ( -55.00) >DroAna_CAF1 24185 114 - 1 CGGGCAUGAAAUGCGGCCGCAUCUGGGGGCCCAUCUGUUGGUGGGCCGGUGACGGUGCUCCCUGCGGUCCGCCGGGAUAGCUGGGCGGGCCAUUGAAUGCCAUGUGGGCAUUGG ..(((.((.....)))))(((...(((((((((((....)))))((((....)))))))))))))((((((((.((....)).))))))))....((((((.....)))))).. ( -60.00) >consensus CGGGCAUAAAAUGCGGACGCAUCUGGGGCACCAUCUGCUGGUGGGCCGGUGACGGUGCACCCUGCGGUCCACCUGGAUAGCUGGGCGGCCCAUUAAAUGCCAUAUGAGCAUUAG ...(((.....)))....((.((...(((.......((.((((.((((....)))).))))..))((.((.(((((....))))).)).)).......)))....))))..... (-25.82 = -26.02 + 0.20)


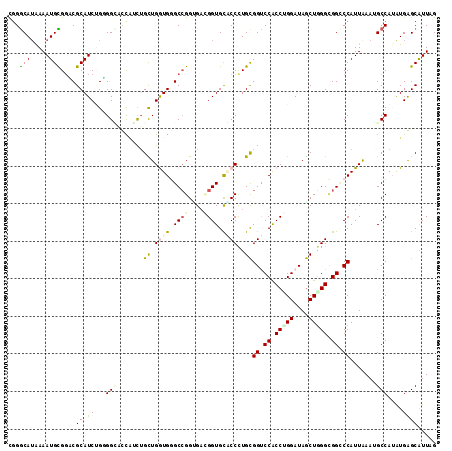
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:17:59 2006