| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,424,514 – 16,424,630 |
| Length | 116 |
| Max. P | 0.671828 |

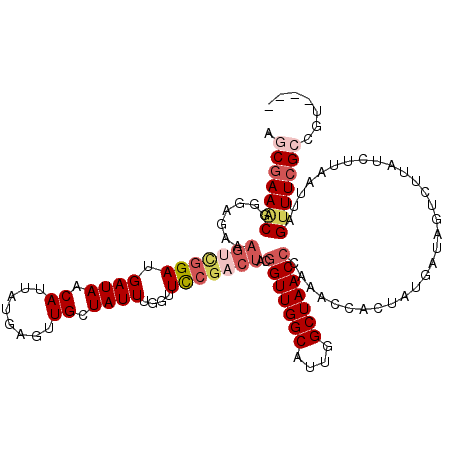
| Location | 16,424,514 – 16,424,630 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.48 |
| Mean single sequence MFE | -27.68 |
| Consensus MFE | -17.64 |
| Energy contribution | -19.56 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.555238 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16424514 116 + 27905053 ----ACGGCGAAACUAAUUAAGAUAAGACUAUCAUAGUGGUUUGGGUUAGCCAAUGCCAACCGUAGUCGGAACCAAAUAGCAACUAAUAAUGUUAUCAUCCGGCUUUCUCCCGCUUCGCU ----..((((((.........((((....))))...((((....((((.((....)).))))..(((((((.....((((((........))))))..))))))).....)))))))))) ( -30.10) >DroSec_CAF1 20260 116 + 1 ----ACGGCGAAACUAAUUAAGAUAAGACUAUCAUUGUGGUUUGGGUUAGCCAAUGCCAACCGUAGUCGAAACCAAAUAGCAACUCAUAAUGUUAUCAUCCGACUUUCGCCCGUUUAGCU ----..(((((((.............((((((.....((((...((....))...))))...))))))........((((((........))))))........)))))))......... ( -26.70) >DroSim_CAF1 20871 115 + 1 ----ACGGCGAAACUAAUUAAGAUAAGACUAUCAUAGUGGUUUG-GUUAGCCAAUGCCAACCGUAGUCGAAACCAAAUAGCAACUCAUAAUGUUAUCAUCCGACUUUCGCCCGUUUCGCU ----..((((((((............((((((.....(((((((-(....)))).))))...))))))((((....((((((........))))))........))))....)))))))) ( -28.70) >DroEre_CAF1 20181 120 + 1 UUUAACUCCGAAACUAAUUAAGGUAAGACUAUCAUAAUGGUUUGGGUUAGCCAAUGCCAACCAUAGUCGGAACCAAAUAACAACUCAUAAUGGUAUCAUCCAACUUUCUUCCGUUUCGUU ........((((((..(..(((((..((((((.....((((...((....))...))))...))))))(((((((...............))))....))).)))))..)..)))))).. ( -25.66) >DroYak_CAF1 22069 99 + 1 UUUAACGCCGAAACG-----CGGGAAAGCUA------UGGUUAGGGUUAGCCAAUGCCAACCGUAGUCGGAUCCAAAUAACAACUCAGAAUGGUAUCAUC----------CCGAUUCGCU ......((.(((...-----(((((..((((------((((...(((........))).))))))))..((((((...............))).))).))----------))).))))). ( -27.26) >consensus ____ACGGCGAAACUAAUUAAGAUAAGACUAUCAUAGUGGUUUGGGUUAGCCAAUGCCAACCGUAGUCGGAACCAAAUAGCAACUCAUAAUGUUAUCAUCCGACUUUCGCCCGUUUCGCU ......((((((((............((((((.....((((...((....))...))))...))))))........((((((........))))))................)))))))) (-17.64 = -19.56 + 1.92)

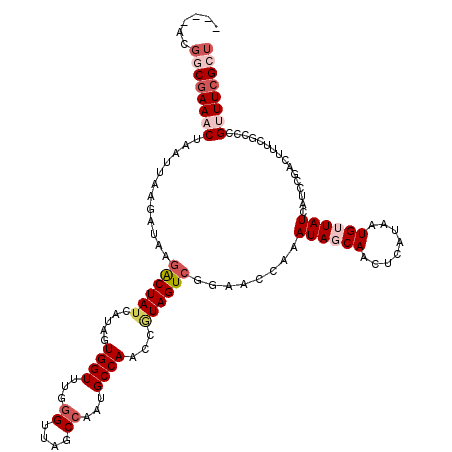
| Location | 16,424,514 – 16,424,630 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.48 |
| Mean single sequence MFE | -32.18 |
| Consensus MFE | -20.90 |
| Energy contribution | -22.62 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.671828 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16424514 116 - 27905053 AGCGAAGCGGGAGAAAGCCGGAUGAUAACAUUAUUAGUUGCUAUUUGGUUCCGACUACGGUUGGCAUUGGCUAACCCAAACCACUAUGAUAGUCUUAUCUUAAUUAGUUUCGCCGU---- .(((((((..((((.(((((((((.((((.......)))).)))))))))..(((((.(((((((....)))))))((........)).)))))...)))).....)))))))...---- ( -35.60) >DroSec_CAF1 20260 116 - 1 AGCUAAACGGGCGAAAGUCGGAUGAUAACAUUAUGAGUUGCUAUUUGGUUUCGACUACGGUUGGCAUUGGCUAACCCAAACCACAAUGAUAGUCUUAUCUUAAUUAGUUUCGCCGU---- .........(((((((.(.....(((((.(((((..((((.....((((((.(.....(((((((....)))))))))))))))))).))))).)))))......).)))))))..---- ( -30.70) >DroSim_CAF1 20871 115 - 1 AGCGAAACGGGCGAAAGUCGGAUGAUAACAUUAUGAGUUGCUAUUUGGUUUCGACUACGGUUGGCAUUGGCUAAC-CAAACCACUAUGAUAGUCUUAUCUUAAUUAGUUUCGCCGU---- .(((((((.(((....)))((((((.(((.......)))((((((((((((.......(((((((....))))))-)))))))....)))))).))))))......)))))))...---- ( -34.81) >DroEre_CAF1 20181 120 - 1 AACGAAACGGAAGAAAGUUGGAUGAUACCAUUAUGAGUUGUUAUUUGGUUCCGACUAUGGUUGGCAUUGGCUAACCCAAACCAUUAUGAUAGUCUUACCUUAAUUAGUUUCGGAGUUAAA ..((((((((((.....(..(((((((.(.......).)))))))..)))))(((((((((((((....)))))))((........))))))))............))))))........ ( -30.30) >DroYak_CAF1 22069 99 - 1 AGCGAAUCGG----------GAUGAUACCAUUCUGAGUUGUUAUUUGGAUCCGACUACGGUUGGCAUUGGCUAACCCUAACCA------UAGCUUUCCCG-----CGUUUCGGCGUUAAA (((((.((((----------((((....)))))))).))))).(((((..((((..(((((((((....))))))).......------..((......)-----))).))))..))))) ( -29.50) >consensus AGCGAAACGGGAGAAAGUCGGAUGAUAACAUUAUGAGUUGCUAUUUGGUUCCGACUACGGUUGGCAUUGGCUAACCCAAACCACUAUGAUAGUCUUAUCUUAAUUAGUUUCGCCGU____ .(((((((.......(((((((.((((.((........)).))))....)))))))..(((((((....)))))))..............................)))))))....... (-20.90 = -22.62 + 1.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:17:53 2006