
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,409,745 – 16,409,886 |
| Length | 141 |
| Max. P | 0.912634 |

| Location | 16,409,745 – 16,409,859 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 82.57 |
| Mean single sequence MFE | -39.80 |
| Consensus MFE | -28.75 |
| Energy contribution | -28.03 |
| Covariance contribution | -0.71 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.762271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
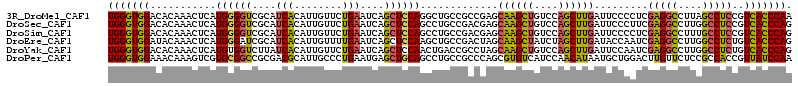
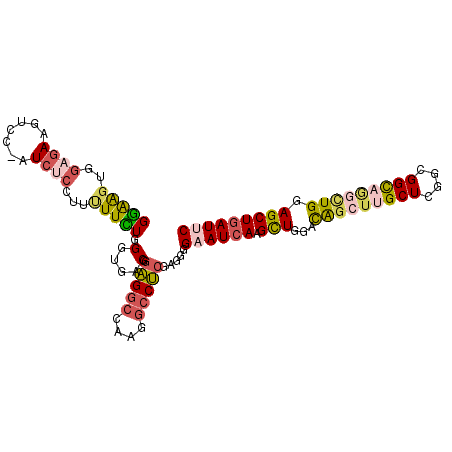
>3R_DroMel_CAF1 16409745 114 + 27905053 UGGGUGGACACAAACUCAUGGGGUCGCAUCACAUUGUUCUGAAUCAGCUCCAGGCUGCCGCCGAGCAAGCUGUCCAGCUUGAUUCCCCUCGAGGCCUUAGCCUCCGUCACCCAA ((((((.((..........((((..(((......)))...(((((.((((..(((....)))))))(((((....)))))))))).))))(((((....))))).)))))))). ( -43.10) >DroSec_CAF1 5385 114 + 1 UGGGUGGACACAAACUCAUGGGGUCGCAUCACAUUGUUCUGAAUCAGCUCCAGCCUGCCGACGAGCAAGCUGUCCAGCUUGAUUCCCUUCGAGGCCUUGGCCUCCGUCACCCAG ((((((.((..........(((...(((......)))...(((((((((.((((.(((......))).))))...))).)))))))))..(((((....))))).)))))))). ( -41.20) >DroSim_CAF1 5389 114 + 1 UGGGUGGACACAAACUCAUGGGGUCGCAUCACAUUGUUCUGAAUCAGCUCCAGCCUGCCGACGAGCAAGCUGUCCAGCUUGAUUCCCCUCGAGGCCUUUGCCUCCGUCACCCAG ((((((.((..........((((..(((......)))...(((((((((.((((.(((......))).))))...))).)))))).))))(((((....))))).)))))))). ( -41.80) >DroEre_CAF1 5444 114 + 1 UGGGUGGAUACAAACUCAUGGGAUCGCAUCACAUUGUUUUGAAUCAGCUCCAAGCUGCCGACUAGCAAGCUAUCUAGCUUGAUACCAAUCGAGGCCUUGGCCUCUGUCACCCAG (((((((...........(((.(((((.(((........)))..((((.....)))).......))(((((....)))))))).)))...(((((....)))))..))))))). ( -36.70) >DroYak_CAF1 5448 114 + 1 UGGGUGGACACAAACUCAUGUGGUCUUAUCACAUUGUUCUGAAUCAGCUCCAACUGACCGCCUAGCAAGCUGUCCAGCUUGAUUCCAAUCGAGGCCUUGGCCUCUGUCACCCAG ((((((.(((.........((((((........(((........)))........))))))....((((((....)))))).........(((((....)))))))))))))). ( -36.99) >DroPer_CAF1 6607 114 + 1 UGGGUGGAAACAAAGUCGUGCGGCCGCGACGCAUUGCCCUGAAUGAGCUGCAGCCUGCCGCCCAGCGUGUCAUCCAACAUAAUGCUGGACUUGUUCUCCGCCACCGUUAUCCAA ..(((((((((((.(((..(((((.((...(((..((.(.....).))))).))..))))).((((((.............)))))))))))))).))))))............ ( -39.02) >consensus UGGGUGGACACAAACUCAUGGGGUCGCAUCACAUUGUUCUGAAUCAGCUCCAGCCUGCCGACGAGCAAGCUGUCCAGCUUGAUUCCCAUCGAGGCCUUGGCCUCCGUCACCCAG (((((((...........((((((....(((........)))....)))))).............((((((....)))))).........(((((....)))))..))))))). (-28.75 = -28.03 + -0.71)
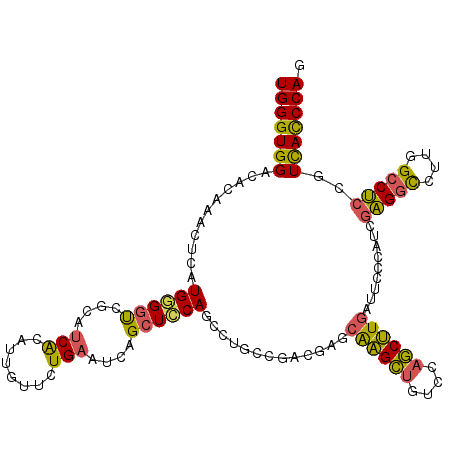
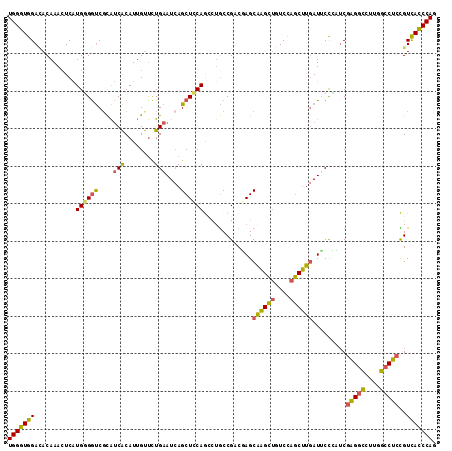
| Location | 16,409,745 – 16,409,859 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 82.57 |
| Mean single sequence MFE | -45.40 |
| Consensus MFE | -33.65 |
| Energy contribution | -35.02 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835765 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
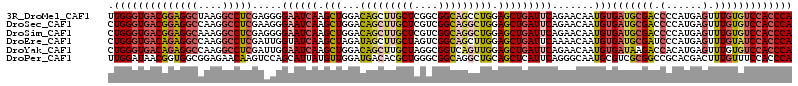
>3R_DroMel_CAF1 16409745 114 - 27905053 UUGGGUGACGGAGGCUAAGGCCUCGAGGGGAAUCAAGCUGGACAGCUUGCUCGGCGGCAGCCUGGAGCUGAUUCAGAACAAUGUGAUGCGACCCCAUGAGUUUGUGUCCACCCA .((((((((((((((....)))))..((((.((((..((((((((((..((.(((....))).))))))).))))).......))))....)))).........))).)))))) ( -47.10) >DroSec_CAF1 5385 114 - 1 CUGGGUGACGGAGGCCAAGGCCUCGAAGGGAAUCAAGCUGGACAGCUUGCUCGUCGGCAGGCUGGAGCUGAUUCAGAACAAUGUGAUGCGACCCCAUGAGUUUGUGUCCACCCA ..(((((((((((((....))))).....((((((.(((...(((((((((....))))))))).))))))))).......)))(((((((.(......).)))))))))))). ( -48.30) >DroSim_CAF1 5389 114 - 1 CUGGGUGACGGAGGCAAAGGCCUCGAGGGGAAUCAAGCUGGACAGCUUGCUCGUCGGCAGGCUGGAGCUGAUUCAGAACAAUGUGAUGCGACCCCAUGAGUUUGUGUCCACCCA ..(((((((((((((....)))))..(((((((((.(((...(((((((((....))))))))).))))))))).......((.....))..))).........))).))))). ( -48.00) >DroEre_CAF1 5444 114 - 1 CUGGGUGACAGAGGCCAAGGCCUCGAUUGGUAUCAAGCUAGAUAGCUUGCUAGUCGGCAGCUUGGAGCUGAUUCAAAACAAUGUGAUGCGAUCCCAUGAGUUUGUAUCCACCCA ..(((((((((((((....)))))((((((...((((((....))))))))))))..((((.....))))...........)))(((((((..........)))))))))))). ( -42.20) >DroYak_CAF1 5448 114 - 1 CUGGGUGACAGAGGCCAAGGCCUCGAUUGGAAUCAAGCUGGACAGCUUGCUAGGCGGUCAGUUGGAGCUGAUUCAGAACAAUGUGAUAAGACCACAUGAGUUUGUGUCCACCCA ..(((((((((((((....))))).....((((((.(((...(((((.(((....))).))))).))))))))).((((.(((((.......)))))..)))).))).))))). ( -44.60) >DroPer_CAF1 6607 114 - 1 UUGGAUAACGGUGGCGGAGAACAAGUCCAGCAUUAUGUUGGAUGACACGCUGGGCGGCAGGCUGCAGCUCAUUCAGGGCAAUGCGUCGCGGCCGCACGACUUUGUUUCCACCCA .........(((((....(((((((((((((....((((....)))).))))))).((.((((((.((.((((......)))).)).))))))))......))))))))))).. ( -42.20) >consensus CUGGGUGACGGAGGCCAAGGCCUCGAGGGGAAUCAAGCUGGACAGCUUGCUCGGCGGCAGGCUGGAGCUGAUUCAGAACAAUGUGAUGCGACCCCAUGAGUUUGUGUCCACCCA .((((((((((((((....))))).....((((((.(((...(((((((((....))))))))).))))))))).......)))(((((((.(......).))))))))))))) (-33.65 = -35.02 + 1.37)
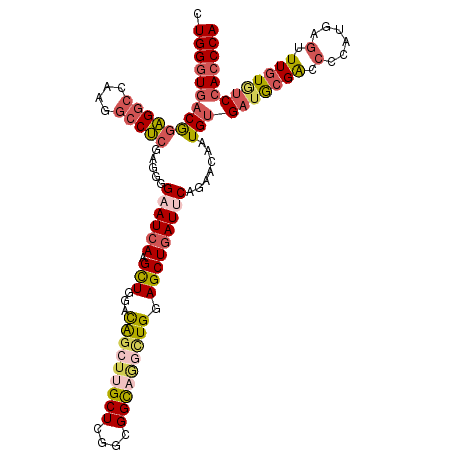
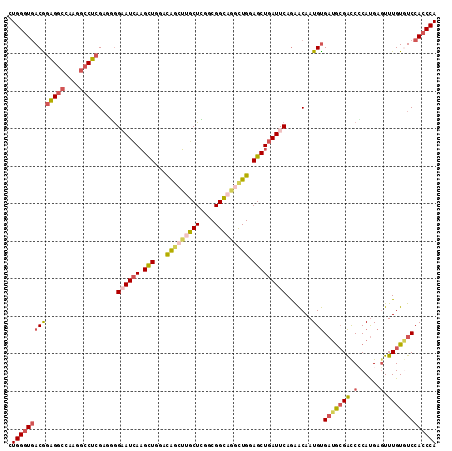
| Location | 16,409,785 – 16,409,886 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 79.14 |
| Mean single sequence MFE | -29.95 |
| Consensus MFE | -16.29 |
| Energy contribution | -16.35 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897062 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16409785 101 + 27905053 GAAUCAGCUCCAGGCUGCCGCCGAGCAAGCUGUCCAGCUUGAUUCCCCUCGAGGCCUUAGCCUCCGUCACCCAAAAAAAUGUGAC-GAACUUCUCCACUUCC (((((.((((..(((....)))))))(((((....)))))))))).....(((((....)))))((((((..........)))))-)............... ( -33.50) >DroSec_CAF1 5425 101 + 1 GAAUCAGCUCCAGCCUGCCGACGAGCAAGCUGUCCAGCUUGAUUCCCUUCGAGGCCUUGGCCUCCGUCACCCAGAAGAAGGAGAU-AGACUUCUCCACUUCC (((((((((.((((.(((......))).))))...))).)))))).....(((((....))))).........((((..(((((.-.....))))).)))). ( -37.30) >DroSim_CAF1 5429 101 + 1 GAAUCAGCUCCAGCCUGCCGACGAGCAAGCUGUCCAGCUUGAUUCCCCUCGAGGCCUUUGCCUCCGUCACCCAGAAGAAGGAGAU-AGACUUCUCCACUUCC (((((((((.((((.(((......))).))))...))).)))))).....(((((....))))).........((((..(((((.-.....))))).)))). ( -35.20) >DroEre_CAF1 5484 101 + 1 GAAUCAGCUCCAAGCUGCCGACUAGCAAGCUAUCUAGCUUGAUACCAAUCGAGGCCUUGGCCUCUGUCACCCAGAAAAACGAGAU-GAACUUCUCCACUUCC ....((((.....))))..(((...((((((....)))))).........(((((....))))).)))............((((.-.....))))....... ( -27.90) >DroYak_CAF1 5488 101 + 1 GAAUCAGCUCCAACUGACCGCCUAGCAAGCUGUCCAGCUUGAUUCCAAUCGAGGCCUUGGCCUCUGUCACCCAGAAAAAGGAGAC-GAACUUCUCCACUUCC (((((((((......(((.((.......)).))).))).)))))).....(((((....)))))...............(((((.-.....)))))...... ( -27.70) >DroPer_CAF1 6647 98 + 1 GAAUGAGCUGCAGCCUGCCGCCCAGCGUGUCAUCCAACAUAAUGCUGGACUUGUUCUCCGCCACCGUUAUCCAAAACA----GAUAGGACUUAUCAACCCUC (((..(((.((.....)).))(((((((.............)))))))..)..)))(((....(.(((......))).----)...)))............. ( -18.12) >consensus GAAUCAGCUCCAGCCUGCCGACGAGCAAGCUGUCCAGCUUGAUUCCCAUCGAGGCCUUGGCCUCCGUCACCCAGAAAAAGGAGAU_GAACUUCUCCACUUCC (((((.((((............))))(((((....)))))))))).....(((((....)))))...................................... (-16.29 = -16.35 + 0.06)

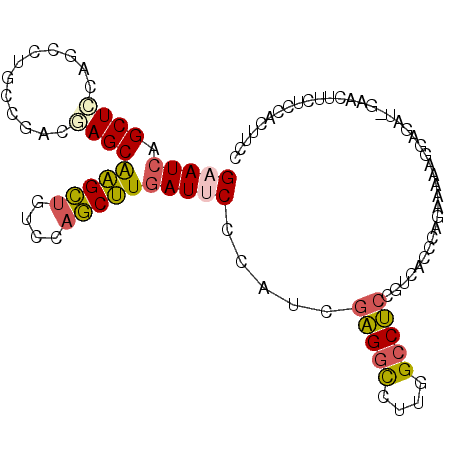
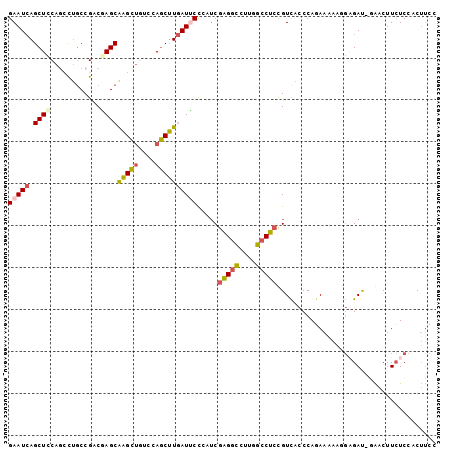
| Location | 16,409,785 – 16,409,886 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 79.14 |
| Mean single sequence MFE | -38.08 |
| Consensus MFE | -25.61 |
| Energy contribution | -26.45 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.912634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16409785 101 - 27905053 GGAAGUGGAGAAGUUC-GUCACAUUUUUUUGGGUGACGGAGGCUAAGGCCUCGAGGGGAAUCAAGCUGGACAGCUUGCUCGGCGGCAGCCUGGAGCUGAUUC ......(((..(((((-(((((..........)))))..(((((...((((((((......((((((....))))))))))).)))))))).)))))..))) ( -35.80) >DroSec_CAF1 5425 101 - 1 GGAAGUGGAGAAGUCU-AUCUCCUUCUUCUGGGUGACGGAGGCCAAGGCCUCGAAGGGAAUCAAGCUGGACAGCUUGCUCGUCGGCAGGCUGGAGCUGAUUC (((((.(((((.....-.)))))..))))).(....).(((((....))))).....((((((.(((...(((((((((....))))))))).))))))))) ( -46.10) >DroSim_CAF1 5429 101 - 1 GGAAGUGGAGAAGUCU-AUCUCCUUCUUCUGGGUGACGGAGGCAAAGGCCUCGAGGGGAAUCAAGCUGGACAGCUUGCUCGUCGGCAGGCUGGAGCUGAUUC (((((.(((((.....-.)))))..))))).(....).(((((....))))).....((((((.(((...(((((((((....))))))))).))))))))) ( -44.60) >DroEre_CAF1 5484 101 - 1 GGAAGUGGAGAAGUUC-AUCUCGUUUUUCUGGGUGACAGAGGCCAAGGCCUCGAUUGGUAUCAAGCUAGAUAGCUUGCUAGUCGGCAGCUUGGAGCUGAUUC (((((..((((.....-.))))...))))).(....).(((((....)))))((((((...((((((....))))))))))))..((((.....)))).... ( -34.40) >DroYak_CAF1 5488 101 - 1 GGAAGUGGAGAAGUUC-GUCUCCUUUUUCUGGGUGACAGAGGCCAAGGCCUCGAUUGGAAUCAAGCUGGACAGCUUGCUAGGCGGUCAGUUGGAGCUGAUUC (((((.(((((.....-.))))).)))))..(....).(((((....))))).....((((((.(((...(((((.(((....))).))))).))))))))) ( -36.70) >DroPer_CAF1 6647 98 - 1 GAGGGUUGAUAAGUCCUAUC----UGUUUUGGAUAACGGUGGCGGAGAACAAGUCCAGCAUUAUGUUGGAUGACACGCUGGGCGGCAGGCUGCAGCUCAUUC ..((((((...((((.((((----(.....)))))...((.((..((.....(((((((.....)))))))......))..)).)).)))).)))))).... ( -30.90) >consensus GGAAGUGGAGAAGUCC_AUCUCCUUUUUCUGGGUGACGGAGGCCAAGGCCUCGAGGGGAAUCAAGCUGGACAGCUUGCUCGGCGGCAGGCUGGAGCUGAUUC (((((..((((.......))))...))))).(....).(((((....))))).....((((((.(((...(((((((((....))))))))).))))))))) (-25.61 = -26.45 + 0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:17:50 2006