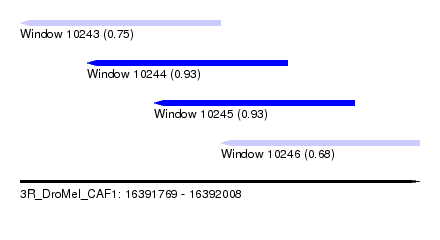
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,391,769 – 16,392,008 |
| Length | 239 |
| Max. P | 0.930477 |

| Location | 16,391,769 – 16,391,889 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -44.30 |
| Consensus MFE | -38.92 |
| Energy contribution | -39.37 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.748718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16391769 120 - 27905053 UACCGAGAGUUGAGAGCUCGACGAGAUGCUUUUGGCUUUUUGGCUUGGUCAGUUGGCUAAUAGUCGAGUCGCGGCUUUCUGGACUGCAUUGGCCGCUAGUAAUCGUAGUUGGCAGUGGUA ((((((((((((.((.((((((.....(((.((((((.........))))))..))).....)))))))).))))))))...(((((...(((..(.(....).)..))).))))))))) ( -42.30) >DroSec_CAF1 12888 120 - 1 UACCGAGAGUUGAGAGCUCGACGAGAUGCUUUUGGCUUUUUGGCUUGGUCAGUUGGCUAAUAGUCGAGUCGCGGCUUUCUGGACUGCAUUGGCCGCUAGCAAUCGUAGUUGGCAGUGCUA ..((((((((((.((.((((((.....(((.((((((.........))))))..))).....)))))))).)))))))).))...((((((.(.((((.......)))).).)))))).. ( -44.00) >DroEre_CAF1 13417 120 - 1 UACCGAGAGCUGAGAGCUCGACGAGAUGCUUUUGGCUUUUUGGCUUGGUCAGUUGGCUAAUAGGCGAGUCGAGGCUUUCUGGACUGCAUUGGCCGCUAGUAAUGGUAGUUGGCAGUGGUA ((((.......(((((((((((....(((((((((((..(((((...)))))..)))))).))))).))))).))))))...(((((...(((..(((....)))..))).))))))))) ( -46.60) >consensus UACCGAGAGUUGAGAGCUCGACGAGAUGCUUUUGGCUUUUUGGCUUGGUCAGUUGGCUAAUAGUCGAGUCGCGGCUUUCUGGACUGCAUUGGCCGCUAGUAAUCGUAGUUGGCAGUGGUA ..((((((((((.((.((((((.....(((.((((((.........))))))..))).....)))))))).)))))))).))(((((...(((..(........)..))).))))).... (-38.92 = -39.37 + 0.45)

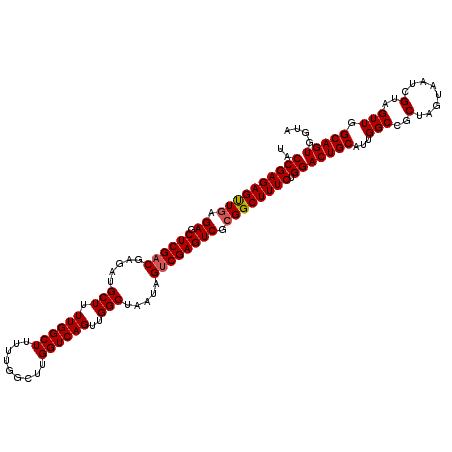
| Location | 16,391,809 – 16,391,929 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -41.33 |
| Consensus MFE | -39.65 |
| Energy contribution | -39.77 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930477 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16391809 120 - 27905053 CGACUGCAAUCGACUUUUUGGCUUGCACAAUUUUUUUGGGUACCGAGAGUUGAGAGCUCGACGAGAUGCUUUUGGCUUUUUGGCUUGGUCAGUUGGCUAAUAGUCGAGUCGCGGCUUUCU .(.((((..((((((..((((((.((.(((.....))).(.(((((((((..(((((((.....)).)))))..)))......))))))).)).)))))).))))))...)))))..... ( -40.70) >DroSec_CAF1 12928 120 - 1 CGACUGCAAUCGACUUUUUGGCUUGCACAAUUUUUUUGGGUACCGAGAGUUGAGAGCUCGACGAGAUGCUUUUGGCUUUUUGGCUUGGUCAGUUGGCUAAUAGUCGAGUCGCGGCUUUCU .(.((((..((((((..((((((.((.(((.....))).(.(((((((((..(((((((.....)).)))))..)))......))))))).)).)))))).))))))...)))))..... ( -40.70) >DroEre_CAF1 13457 119 - 1 CGACUGCAAUCGACUUUUUGGCUUGCACAAUUU-UUUGGGUACCGAGAGCUGAGAGCUCGACGAGAUGCUUUUGGCUUUUUGGCUUGGUCAGUUGGCUAAUAGGCGAGUCGAGGCUUUCU .((..((..(((((((...(.((.((.(((((.-.(..(((...((((((..(((((((.....)).)))))..))))))..)))..)..)))))))....)).)))))))).))..)). ( -42.60) >consensus CGACUGCAAUCGACUUUUUGGCUUGCACAAUUUUUUUGGGUACCGAGAGUUGAGAGCUCGACGAGAUGCUUUUGGCUUUUUGGCUUGGUCAGUUGGCUAAUAGUCGAGUCGCGGCUUUCU .((..((...((((((...((((.((.(((((...(..(((...((((((..(((((((.....)).)))))..))))))..)))..)..)))))))....))))))))))..))..)). (-39.65 = -39.77 + 0.11)


| Location | 16,391,849 – 16,391,969 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -41.37 |
| Consensus MFE | -41.67 |
| Energy contribution | -41.23 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.02 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927051 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16391849 120 - 27905053 GCCAGGAUUGUGCAAUCCGCAAUCGGCAAUCCGCAGCUGACGACUGCAAUCGACUUUUUGGCUUGCACAAUUUUUUUGGGUACCGAGAGUUGAGAGCUCGACGAGAUGCUUUUGGCUUUU .((((((((((((((.((....(((((........)))))(((......))).......)).))))))))))...)))).....((((((..(((((((.....)).)))))..)))))) ( -40.50) >DroSec_CAF1 12968 120 - 1 GCCAGGAUUGUGCAAUCCGCAAUCGGCAAUCCGCAGCUGACGACUGCAAUCGACUUUUUGGCUUGCACAAUUUUUUUGGGUACCGAGAGUUGAGAGCUCGACGAGAUGCUUUUGGCUUUU .((((((((((((((.((....(((((........)))))(((......))).......)).))))))))))...)))).....((((((..(((((((.....)).)))))..)))))) ( -40.50) >DroEre_CAF1 13497 119 - 1 GCCAGGAUUGUGCAAUCCGCAAUCAGCAAUCCGCAGCUGACGACUGCAAUCGACUUUUUGGCUUGCACAAUUU-UUUGGGUACCGAGAGCUGAGAGCUCGACGAGAUGCUUUUGGCUUUU .((((((((((((((.((....(((((........)))))(((......))).......)).)))))))))).-.)))).....((((((..(((((((.....)).)))))..)))))) ( -43.10) >consensus GCCAGGAUUGUGCAAUCCGCAAUCGGCAAUCCGCAGCUGACGACUGCAAUCGACUUUUUGGCUUGCACAAUUUUUUUGGGUACCGAGAGUUGAGAGCUCGACGAGAUGCUUUUGGCUUUU .((((((((((((((.((....(((((........)))))(((......))).......)).))))))))))...)))).....((((((..(((((((.....)).)))))..)))))) (-41.67 = -41.23 + -0.44)

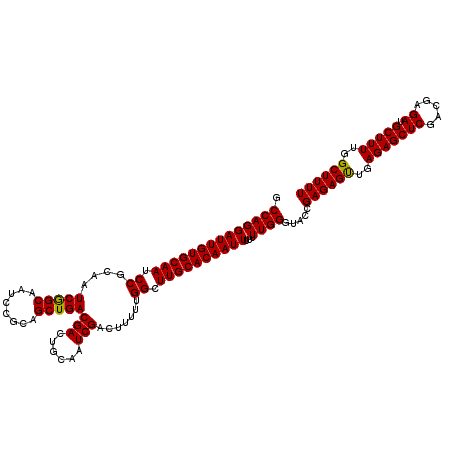
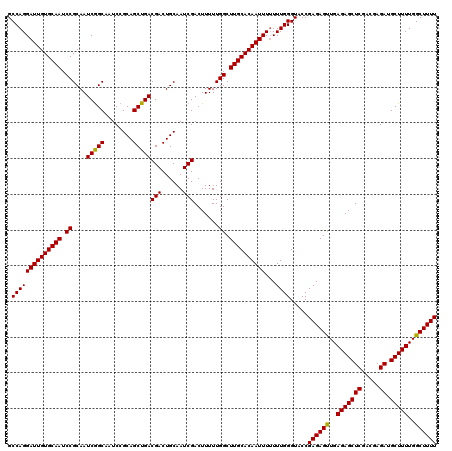
| Location | 16,391,889 – 16,392,008 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.04 |
| Mean single sequence MFE | -36.07 |
| Consensus MFE | -33.80 |
| Energy contribution | -34.13 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.677172 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16391889 119 - 27905053 CACACACACAC-AGCAGCAGCCAACGAACAGCAACAGGUUGCCAGGAUUGUGCAAUCCGCAAUCGGCAAUCCGCAGCUGACGACUGCAAUCGACUUUUUGGCUUGCACAAUUUUUUUGGG ...........-....((((((((....((((..(.(((((((..(((((((.....)))))))))))))).)..)))).(((......))).....))))).))).(((.....))).. ( -35.40) >DroSec_CAF1 13008 112 - 1 CACACAC--------AGCAGCCAACGAACAGCAACAGGUUGCCAGGAUUGUGCAAUCCGCAAUCGGCAAUCCGCAGCUGACGACUGCAAUCGACUUUUUGGCUUGCACAAUUUUUUUGGG .......--------.((((((((....((((..(.(((((((..(((((((.....)))))))))))))).)..)))).(((......))).....))))).))).(((.....))).. ( -35.40) >DroEre_CAF1 13537 119 - 1 CACACACGCGCGUGCAGCAGCCAACCAACAGCAACAGGUUGCCAGGAUUGUGCAAUCCGCAAUCAGCAAUCCGCAGCUGACGACUGCAAUCGACUUUUUGGCUUGCACAAUUU-UUUGGG ...........(((((..((((((....((((..(.((((((...(((((((.....))))))).)))))).)..)))).(((......))).....))))))))))).....-...... ( -37.40) >consensus CACACAC_C_C__GCAGCAGCCAACGAACAGCAACAGGUUGCCAGGAUUGUGCAAUCCGCAAUCGGCAAUCCGCAGCUGACGACUGCAAUCGACUUUUUGGCUUGCACAAUUUUUUUGGG ................((((((((....((((..(.(((((((..(((((((.....)))))))))))))).)..)))).(((......))).....))))).))).(((.....))).. (-33.80 = -34.13 + 0.33)

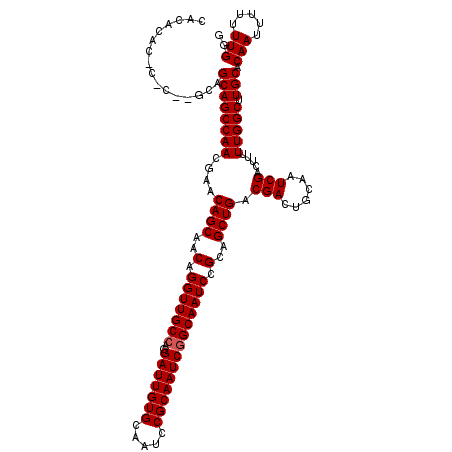
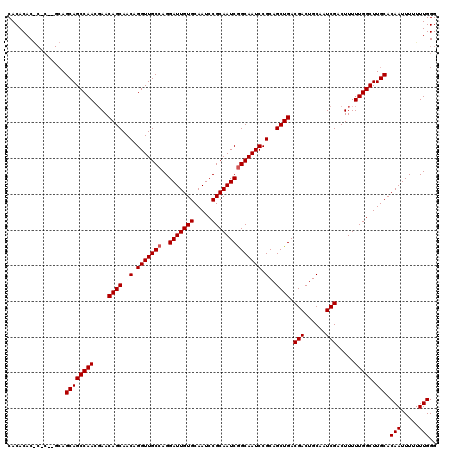
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:17:35 2006