
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,379,939 – 16,380,154 |
| Length | 215 |
| Max. P | 0.972549 |

| Location | 16,379,939 – 16,380,038 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 95.29 |
| Mean single sequence MFE | -23.60 |
| Consensus MFE | -22.42 |
| Energy contribution | -22.53 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16379939 99 + 27905053 GAAAAUGAAUAGAAAAUCUCAAUAUGCAUUCAUACCCGAAUGCGCGACUAUGGAUAAAUAAACGGAGGGACAUAGGAAACUUUGACACUCCACUACGCU .........................((((((......))))))(((..(((......)))...((((...((.((....)).))...))))....))). ( -22.20) >DroSec_CAF1 1298 99 + 1 GAAAAUAAAUGGAAAAUCUCAAUAUGCAUUCAUACCCGAAUGCGCGACUAUGGAUCAAUAAACGGAGGGACAUAGGAAACUUUGACUCUCCACUACACU .........(((...((((.....(((((((......))))))).......))))........((((((.((.((....)).)).)))))).))).... ( -23.30) >DroSim_CAF1 1294 99 + 1 GAAAAUGAAUAGUAAAUCUCAAUAUGCAUUCAUACCCGAAUGCGCGACUAUGGAUAAACAAACGGAGGGACAUAGGAAACUUUGACUCUCCACUACACU .........((((..((((.....(((((((......))))))).......))))........((((((.((.((....)).)).)))))))))).... ( -25.30) >consensus GAAAAUGAAUAGAAAAUCUCAAUAUGCAUUCAUACCCGAAUGCGCGACUAUGGAUAAAUAAACGGAGGGACAUAGGAAACUUUGACUCUCCACUACACU .........(((...((((.....(((((((......))))))).......))))........((((((.((.((....)).)).)))))).))).... (-22.42 = -22.53 + 0.11)

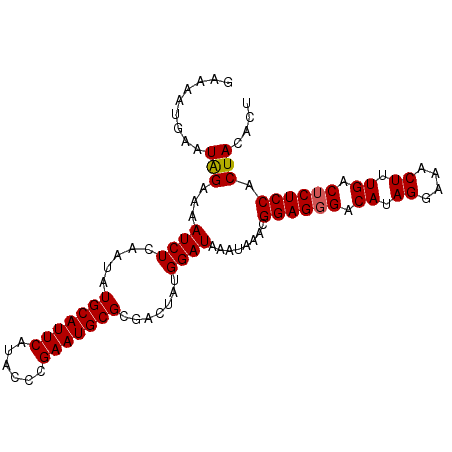

| Location | 16,379,968 – 16,380,078 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 95.76 |
| Mean single sequence MFE | -25.83 |
| Consensus MFE | -25.63 |
| Energy contribution | -25.30 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.786691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16379968 110 + 27905053 UCAUACCCGAAUGCGCGACUAUGGAUAAAUAAACGGAGGGACAUAGGAAACUUUGACACUCCACUACGCUCUAUAAUUUAGUUCGGUUUGAAGUCAAAUACUCGACAUUU (((...((((((....((.((((((.........((((...((.((....)).))...)))).......)))))).))..))))))..))).(((........))).... ( -24.19) >DroSec_CAF1 1327 110 + 1 UCAUACCCGAAUGCGCGACUAUGGAUCAAUAAACGGAGGGACAUAGGAAACUUUGACUCUCCACUACACUCUAUAAUUUAGUUUGGUUUGCAGUCAAAUACUCGACAUUU .......(((.((...((((..(((((((.....((((((.((.((....)).)).))))))........(((.....))).)))))))..))))...)).)))...... ( -26.40) >DroSim_CAF1 1323 110 + 1 UCAUACCCGAAUGCGCGACUAUGGAUAAACAAACGGAGGGACAUAGGAAACUUUGACUCUCCACUACACUCUAAAAUUUAGUUUGGUUUGCAGUCAAAUACUCGACAUUU ........(((((..(((.(((.(((...(((((((((((.((.((....)).)).))))))....((..(((.....)))..)))))))..)))..))).))).))))) ( -26.90) >consensus UCAUACCCGAAUGCGCGACUAUGGAUAAAUAAACGGAGGGACAUAGGAAACUUUGACUCUCCACUACACUCUAUAAUUUAGUUUGGUUUGCAGUCAAAUACUCGACAUUU ........(((((..(((.(((.(((...(((((((((((.((.((....)).)).))))))....((..(((.....)))..)))))))..)))..))).))).))))) (-25.63 = -25.30 + -0.33)

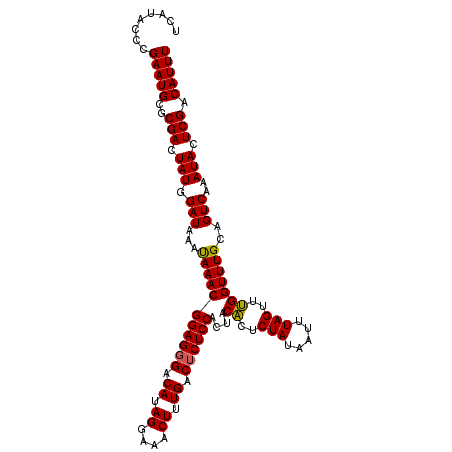
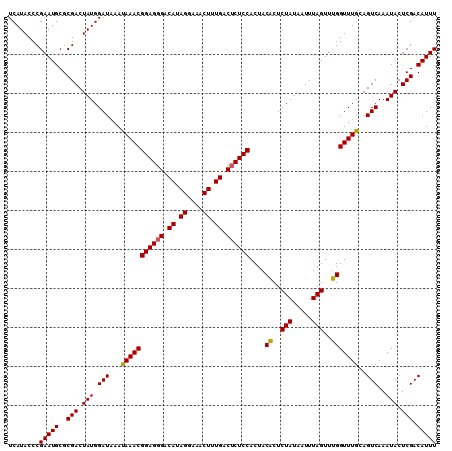
| Location | 16,379,998 – 16,380,118 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -29.37 |
| Consensus MFE | -26.17 |
| Energy contribution | -26.73 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931661 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16379998 120 + 27905053 AAACGGAGGGACAUAGGAAACUUUGACACUCCACUACGCUCUAUAAUUUAGUUCGGUUUGAAGUCAAAUACUCGACAUUUUACUGUCGACAAACACAAAAAUUCAAGAGUUUUCAUUGUG ....((((...((.((....)).))...))))....((..(((.....)))..))(((((....)))))..((((((......))))))....((((((((((....)))))...))))) ( -26.10) >DroSec_CAF1 1357 120 + 1 AAACGGAGGGACAUAGGAAACUUUGACUCUCCACUACACUCUAUAAUUUAGUUUGGUUUGCAGUCAAAUACUCGACAUUUUACUGCCGACAAACACAAAAAUUCAAGAGUUUUCAUUGUG ....((((((.((.((....)).)).)))))).....(((((..(((((.(((((.((.(((((.((((.......)))).))))).)))))))....)))))..))))).......... ( -30.60) >DroSim_CAF1 1353 120 + 1 AAACGGAGGGACAUAGGAAACUUUGACUCUCCACUACACUCUAAAAUUUAGUUUGGUUUGCAGUCAAAUACUCGACAUUUUACUGCCGACAAACACAAAAAUUCAAGAGUUUUCAUUGUA ....((((((.((.((....)).)).)))))).....(((((..(((((.(((((.((.(((((.((((.......)))).))))).)))))))....)))))..))))).......... ( -31.40) >consensus AAACGGAGGGACAUAGGAAACUUUGACUCUCCACUACACUCUAUAAUUUAGUUUGGUUUGCAGUCAAAUACUCGACAUUUUACUGCCGACAAACACAAAAAUUCAAGAGUUUUCAUUGUG ....((((((.((.((....)).)).)))))).....(((((..(((((.(((((.((.(((((.((((.......)))).))))).)))))))....)))))..))))).......... (-26.17 = -26.73 + 0.56)



| Location | 16,380,038 – 16,380,154 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 95.40 |
| Mean single sequence MFE | -26.37 |
| Consensus MFE | -24.31 |
| Energy contribution | -23.87 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931553 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16380038 116 - 27905053 AACAAAAAUUAGGGAAACGCUAAAUUACAUUGGCGACACAAUGAAAACUCUUGAAUUUUUGUGUUUGUCGACAGUAAAAUGUCGAGUAUUUGACUUCAAACCGAACUAAAUUAUAG .(((((((((..(((..((((((......)))))).(.....).....)))..)))))))))(((((((((((......))))))...((((....)))).))))).......... ( -25.80) >DroSec_CAF1 1397 116 - 1 AACAAAAAUUAGGGGAACGCUAAAUUAUAUUGGCGACACAAUGAAAACUCUUGAAUUUUUGUGUUUGUCGGCAGUAAAAUGUCGAGUAUUUGACUGCAAACCAAACUAAAUUAUAG .((((((((((((((..((((((......)))))).(.....)....)))))).))))))))(((((...(((((.(((((.....))))).)))))....))))).......... ( -27.90) >DroSim_CAF1 1393 116 - 1 AACAAAAAUUAGGAAAACGCUAAAUUAUAUUGGCGAUACAAUGAAAACUCUUGAAUUUUUGUGUUUGUCGGCAGUAAAAUGUCGAGUAUUUGACUGCAAACCAAACUAAAUUUUAG .(((((((((.......((((((......))))))...(((.((....))))))))))))))(((((...(((((.(((((.....))))).)))))....))))).......... ( -25.40) >consensus AACAAAAAUUAGGGAAACGCUAAAUUAUAUUGGCGACACAAUGAAAACUCUUGAAUUUUUGUGUUUGUCGGCAGUAAAAUGUCGAGUAUUUGACUGCAAACCAAACUAAAUUAUAG .(((((((((.......((((((......))))))...(((.((....))))))))))))))(((((((((((......))))))...((((....)))).))))).......... (-24.31 = -23.87 + -0.44)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:17:22 2006