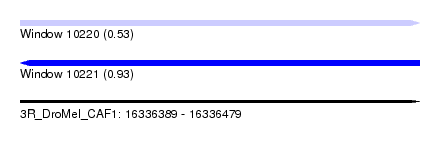
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,336,389 – 16,336,479 |
| Length | 90 |
| Max. P | 0.929276 |

| Location | 16,336,389 – 16,336,479 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 81.71 |
| Mean single sequence MFE | -24.98 |
| Consensus MFE | -13.56 |
| Energy contribution | -15.07 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.54 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.530077 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
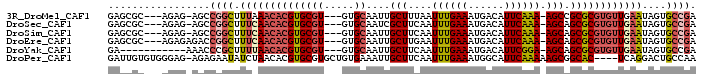
>3R_DroMel_CAF1 16336389 90 + 27905053 UCGGCACUAUUCAACACGCGCGGCU-UUUGAAUGUCAUUUCAAAUUAAAGCAAUUGCAC---ACGCACGUGUUUAAAGCCGGCU-CUCU---GCGCUC .((((..((...((((((.(((...-((((((......)))))).....((....))..---.))).))))))))..))))((.-....---)).... ( -25.10) >DroSec_CAF1 38855 90 + 1 UCGGCACUAUUCAACACGCGCUGCU-UUUGAAUGUCAUUUCAAAUUGAAGCGAUUGCAC---ACGCACGUGUUGAAAGCCGGCU-CUCU---GCGCUC .((((....(((((((((((((...-((((((......))))))....))))..(((..---..)))))))))))).))))((.-....---)).... ( -30.90) >DroSim_CAF1 41404 90 + 1 UCGGCACUAUUCAACACGCGCUGCU-UUUGAAUGUCAUUUCAAAUUGAAGCAAUUGCAC---ACGCACGUGUUGAAAGCCGGCU-CUCU---GCGCUC .((((....(((((((((.(((((.-.(((....(((........)))..)))..))).---..)).))))))))).))))((.-....---)).... ( -30.20) >DroEre_CAF1 38019 91 + 1 UCGGCACUAUUCAACACGCGCUGCU-UUUGAAUGUCAUUUCAAAUUCAAGCAAUUGCAC---ACGCACGUGUUGAAAGCCGGUCUCUCU---GCGCUC (((((....(((((((((.((((..-((((((......))))))..)).((....))..---..)).))))))))).))))).......---...... ( -28.70) >DroYak_CAF1 55460 83 + 1 UCGGCACUAUUCAACACGCGCUGCU-UCCGAAUGUCAUUUCAAAUUGAAGCAAUUGCAC---ACGCACGUGUUAAAAGCGGGUUU-----------UC ............(((.(((((((((-((....((......))....))))))...))((---((....)))).....))).))).-----------.. ( -16.30) >DroPer_CAF1 64238 93 + 1 UUGGCAGUCCUGA----GUGCCGCUUUUUGAAUGCCAUUUCAAAUUGAAGCAAUUUCACAGCACGCACGUGUUAGAUAUUCUCU-CUCCCACACAAUC .(((.((....((----(((.(((((((((((......)))))...)))))........(((((....))))).).)))))...-)).)))....... ( -18.70) >consensus UCGGCACUAUUCAACACGCGCUGCU_UUUGAAUGUCAUUUCAAAUUGAAGCAAUUGCAC___ACGCACGUGUUGAAAGCCGGCU_CUCU___GCGCUC .((((.......((((((.((((((.((((((......))))))....))))............)).))))))....))))................. (-13.56 = -15.07 + 1.50)

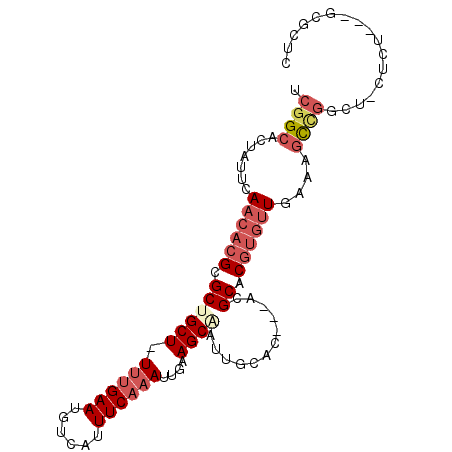

| Location | 16,336,389 – 16,336,479 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 81.71 |
| Mean single sequence MFE | -29.98 |
| Consensus MFE | -23.66 |
| Energy contribution | -24.88 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929276 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
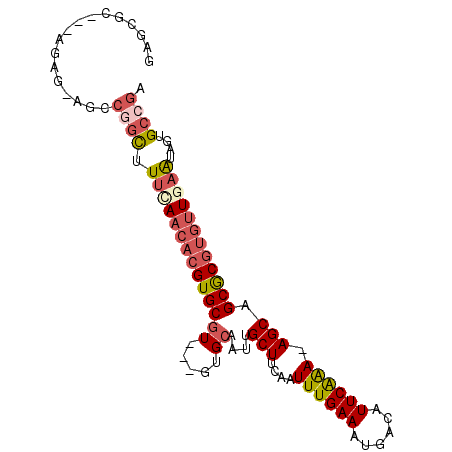
>3R_DroMel_CAF1 16336389 90 - 27905053 GAGCGC---AGAG-AGCCGGCUUUAAACACGUGCGU---GUGCAAUUGCUUUAAUUUGAAAUGACAUUCAAA-AGCCGCGCGUGUUGAAUAGUGCCGA ..((..---....-.))((((....((((((((((.---((((....)).....((((((......))))))-.)))))))))))).......)))). ( -29.10) >DroSec_CAF1 38855 90 - 1 GAGCGC---AGAG-AGCCGGCUUUCAACACGUGCGU---GUGCAAUCGCUUCAAUUUGAAAUGACAUUCAAA-AGCAGCGCGUGUUGAAUAGUGCCGA ..((..---....-.))((((.((((((((((((.(---((((....)).....((((((......))))))-.)))))))))))))))....)))). ( -33.10) >DroSim_CAF1 41404 90 - 1 GAGCGC---AGAG-AGCCGGCUUUCAACACGUGCGU---GUGCAAUUGCUUCAAUUUGAAAUGACAUUCAAA-AGCAGCGCGUGUUGAAUAGUGCCGA ..((..---....-.))((((.((((((((((((.(---((((....)).....((((((......))))))-.)))))))))))))))....)))). ( -33.40) >DroEre_CAF1 38019 91 - 1 GAGCGC---AGAGAGACCGGCUUUCAACACGUGCGU---GUGCAAUUGCUUGAAUUUGAAAUGACAUUCAAA-AGCAGCGCGUGUUGAAUAGUGCCGA ......---........((((.((((((((((((.(---((((....)).....((((((......))))))-.)))))))))))))))....)))). ( -32.70) >DroYak_CAF1 55460 83 - 1 GA-----------AAACCCGCUUUUAACACGUGCGU---GUGCAAUUGCUUCAAUUUGAAAUGACAUUCGGA-AGCAGCGCGUGUUGAAUAGUGCCGA ..-----------.....(((((((((((((((((.---...)...((((((....((......))....))-)))))))))))))))).)))).... ( -28.00) >DroPer_CAF1 64238 93 - 1 GAUUGUGUGGGAG-AGAGAAUAUCUAACACGUGCGUGCUGUGAAAUUGCUUCAAUUUGAAAUGGCAUUCAAAAAGCGGCAC----UCAGGACUGCCAA ......(((..((-(.......)))..)))((((.((((.((((..((((((.....))...))))))))...))))))))----...((....)).. ( -23.60) >consensus GAGCGC___AGAG_AGCCGGCUUUCAACACGUGCGU___GUGCAAUUGCUUCAAUUUGAAAUGACAUUCAAA_AGCAGCGCGUGUUGAAUAGUGCCGA .................((((.((((((((((((((.....))....(((....((((((......)))))).))).))))))))))))....)))). (-23.66 = -24.88 + 1.23)
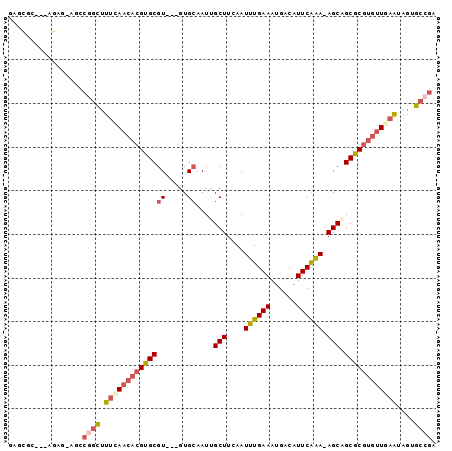
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:17:11 2006