| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,334,278 – 16,334,395 |
| Length | 117 |
| Max. P | 0.921086 |

| Location | 16,334,278 – 16,334,395 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.81 |
| Mean single sequence MFE | -34.45 |
| Consensus MFE | -20.78 |
| Energy contribution | -20.12 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921086 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
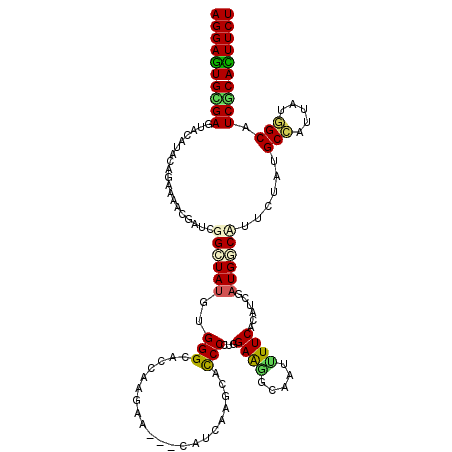
>3R_DroMel_CAF1 16334278 117 - 27905053 AGGAAUGCGACUACAUCCAGAAGACCAUCGGGUAUGUGGGCUCUAAGCA---CGUCAAGCACCCCUGGAGGGCCAUCUUCACGUCGAUGCCAUUUUAUGCCAUCAUGGCCUCGCAUUUCU ((((((((((.....(((((..(.....)((((.((((.((.....)).---)).))...))))))))).((((((.........((((.(((...))).)))))))))))))))))))) ( -36.30) >DroVir_CAF1 63990 117 - 1 AGGAGUGUGAGUACAUACAGAAAUCGAUUGGCUAUGUGGGCGCCAAGAA---GAUCAAGCACCCUUGGAAAGCGUUUUUCACAUCGAUGCCCUUCUAUGCCAUUAUUGCAUCACACUUCU (((((((((((((((((.((..(.....)..)).))))((((...((((---(.(((((....)))))...(((((.........))))).))))).)))).....))).)))))))))) ( -31.90) >DroGri_CAF1 52009 120 - 1 AGGAGUGCGAGUACAUACAUAAAACGAUUGGCUAUACGGGUUCAAACACCAAUAUCAAGCACCCGUGGAAGGCCAUAUUCACCUCAAUGGCCUUUUAUGCAAUUAUUGCAUCGCAUUUCU ((((((((((((....)).......(((((((((((..(((......)))..)))..)))...(((((((((((((..........))))))))))))))))))......)))))))))) ( -36.20) >DroWil_CAF1 54406 117 - 1 AGGAAUGCGAUUACAUACAGAAGACAAUCGGCUAUGUGGGCACCAAGCA---UAUUAAGCAUCCAUGGAAGUCAAUUUUCACAUCGAUGGCAUUCUAUGCCAUUAUGGCUUCACAUUUCU .(((.(((..(((((((..((......))...)))))))((.....)).---......))))))((((((((((...........((((((((...)))))))).))))))).))).... ( -28.10) >DroMoj_CAF1 46856 117 - 1 AGGAGUGCGAGUACAUACAGAAAACGAUUGGAUACACGGGCCCCCAGAC---CAUCAAGCACCCGUGGAAGGCAAUCUUCACAUCCAUGGCUUUCUAUGCAAUCAUUGCCUCGCACUUCU (((((((((((.......(((((.(.((.((((.((((((.........---.........))))))((((.....))))..)))))).).)))))..(((.....)))))))))))))) ( -38.97) >DroAna_CAF1 28737 117 - 1 AGGAGUGUGAAUACAUUCAGAAGACCAUCGGCUAUGUGGGCACAAAACA---CAUCAAGCAUCCCUGGAAGUCAAUUUUCACAUCCAUGGCUUUCUACGCCAUUAUGGCUUCGCACUUCU (((((((((((.......((((..((((..((((((((.........))---)))..)))......(((.((........)).)))))))..))))..(((.....)))))))))))))) ( -35.20) >consensus AGGAGUGCGAGUACAUACAGAAAACGAUCGGCUAUGUGGGCACCAAGAA___CAUCAAGCACCCCUGGAAGGCAAUUUUCACAUCGAUGGCAUUCUAUGCCAUUAUGGCAUCGCACUUCU ((((((((((...................((((((..(((.....................)))...((((.....))))......))))))......(((.....))).)))))))))) (-20.78 = -20.12 + -0.66)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:17:07 2006