
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,331,426 – 16,331,614 |
| Length | 188 |
| Max. P | 0.997152 |

| Location | 16,331,426 – 16,331,545 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 93.87 |
| Mean single sequence MFE | -36.26 |
| Consensus MFE | -31.80 |
| Energy contribution | -31.96 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
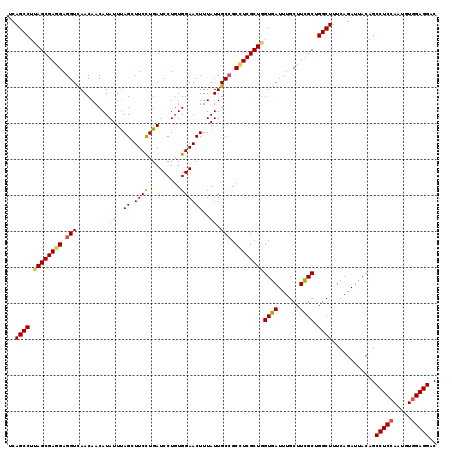
>3R_DroMel_CAF1 16331426 119 + 27905053 UCAGCCUCAGCGAGGAGGUCAACAACAUAUUCAGCUUCCUGAUCCUGUGGAACUUUAUUGCCGCAUCGCUCGUGAUUUGCUUCGCUGGCUUUCAGAUUACAGCCUCAAAUGUGGAGGAC ....(((((((((((((((((.........((((....))))...(((((..........))))).......)))))).)))))))((((..........)))).........)))).. ( -31.10) >DroSec_CAF1 34635 117 + 1 --AGCCUUAGCGAGGAGGUCAACAACAUAUUUAGUUUCUUGAUCCUGUGGAACUUUAUUGCCGCCUCGCUGGUGAUUUGCUUUGCAGGCUUUCAGAUUACAGCCUCCAAUGUGGAGGAC --(((((((((((((.((.((.....(((...((((((..........)))))).))))))).))))))))(..(......)..))))))............(((((.....))))).. ( -36.20) >DroSim_CAF1 36240 119 + 1 UCAGCCUCAGCGAGGAGGUCAACAACAUAUUUAGUUUCCUGAUCCUGUGGAACUUUAUUGCAGCCUCGCUGGUGAUUUGCUUUGCAGGCUUUCAGAUUACAGCCUCCAAUGUGGAGGAC ..(((((((((((((.(((((..(((.......)))...)))))(((..(.......)..)))))))))))(..(......)..))))))............(((((.....))))).. ( -38.00) >DroEre_CAF1 32806 119 + 1 UCAGCCUUAGCGAGGAGGUUAACAACAUAUUUAGCUUCCUGAUCCUGUGGAACUUUAUUGCCGCUUCGCUGGUGAUUUGCUUCGCUGGCUUUCAGAUUACAGCCUCCAAUGUGGAGGAC ..((((..((((((((((((((........))))))))).......((((..........)))).)))))(((((......)))))))))............(((((.....))))).. ( -37.90) >DroYak_CAF1 47842 119 + 1 UCAGCCUUAGCGAGGAGGUUAACAACAUAUUUAGCUUCCUGAUCCUCUGGAACUUUAUUGCCGCCUCGCUGGUGAUUUGCUUCGCUGGCUUUCAGAUAACAGCCUCCAAUGUGGAGGAC ..((((..(((((((.(((.........((..((.((((.(.....).))))))..)).))).)))))))(((((......)))))))))............(((((.....))))).. ( -38.10) >consensus UCAGCCUUAGCGAGGAGGUCAACAACAUAUUUAGCUUCCUGAUCCUGUGGAACUUUAUUGCCGCCUCGCUGGUGAUUUGCUUCGCUGGCUUUCAGAUUACAGCCUCCAAUGUGGAGGAC ..((((.((((((((.(((.........((..((.((((.........))))))..)).))).))))))))((((......)))).))))............(((((.....))))).. (-31.80 = -31.96 + 0.16)


| Location | 16,331,505 – 16,331,614 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 93.58 |
| Mean single sequence MFE | -30.17 |
| Consensus MFE | -29.60 |
| Energy contribution | -29.52 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.81 |
| SVM RNA-class probability | 0.997152 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
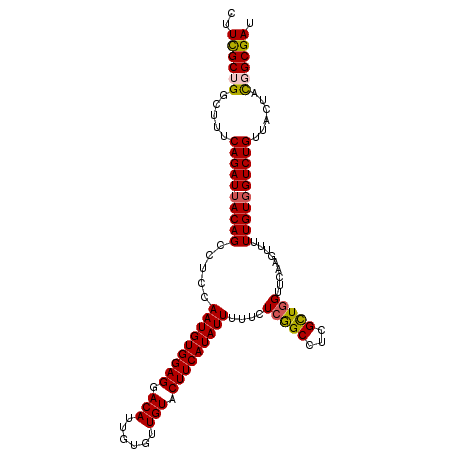
>3R_DroMel_CAF1 16331505 109 + 27905053 CUUCGCUGGCUUUCAGAUUACAGCCUCAAAUGUGGAGGACAUAGUGUUGUACUUCAUAUUUUUUUCAGCUUCGCUGGUUCAAGUCUUUGUGGUCUGUUACUACGGCGAU ..((((((.....((((((((((....((((((((((.(((......))).))))))))))...(((((...))))).........))))))))))......)))))). ( -35.50) >DroSec_CAF1 34712 109 + 1 CUUUGCAGGCUUUCAGAUUACAGCCUCCAAUGUGGAGGACAUUGUUUUGUACUUCAUAUUUUUCUCAGCUUCGCUGGUUCAAGUCUUUGUGGUCUGUUACUACGGCGAU ..((((.......((((((((((.....(((((((((.(((......))).)))))))))....(((((...))))).........))))))))))........)))). ( -28.56) >DroSim_CAF1 36319 109 + 1 CUUUGCAGGCUUUCAGAUUACAGCCUCCAAUGUGGAGGACAUAGUGUUGUACUUCAUAUUUUUCUCGGCCUCGCUGGUUCAAGUUUUUGUGGUCUGUUACUACGGCGAU ..((((.......((((((((((((...(((((((((.(((......))).)))))))))......)))...(((......)))...)))))))))........)))). ( -28.36) >DroEre_CAF1 32885 109 + 1 CUUCGCUGGCUUUCAGAUUACAGCCUCCAAUGUGGAGGACAUUGUGUUGUACUUCAUAUUUUUCUCGGCCUCGCUGGUACAAGUUUUUGUGGUCUGUUACUAUGGCGAU ..((((((.....(((((((((((((((.....)))))........((((((.............((((...))))))))))....))))))))))......)))))). ( -32.11) >DroYak_CAF1 47921 109 + 1 CUUCGCUGGCUUUCAGAUAACAGCCUCCAAUGUGGAGGACAUUGUGUUGUACUUCAUAUUUUUCUCGGCCUCGUUAGUUCAAGUUUUUGUGGUCUGUUACUAUGGCGAU ..((((((.....(((((.((((((...(((((((((.(((......))).)))))))))......)))...(......).......))).)))))......)))))). ( -26.30) >consensus CUUCGCUGGCUUUCAGAUUACAGCCUCCAAUGUGGAGGACAUUGUGUUGUACUUCAUAUUUUUCUCGGCCUCGCUGGUUCAAGUUUUUGUGGUCUGUUACUACGGCGAU ..((((((.....((((((((((.....(((((((((.(((......))).)))))))))....(((((...))))).........))))))))))......)))))). (-29.60 = -29.52 + -0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:17:04 2006