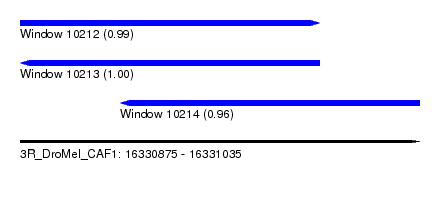
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,330,875 – 16,331,035 |
| Length | 160 |
| Max. P | 0.999314 |

| Location | 16,330,875 – 16,330,995 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.42 |
| Mean single sequence MFE | -32.82 |
| Consensus MFE | -28.26 |
| Energy contribution | -28.82 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989267 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16330875 120 + 27905053 GAUAUUUCCUUUAGAUUUAGAAGCGCAGCGGAAGUAUAACGUAUCGUUUUACCGGAAACACAUGAACAGGGUCAUGACCCUAUUCACCAUCCUCUGCAUGACCUACACCUCGUCAUUUAG ........................((((.(((.(((.((((...)))).))).(....)...((((.(((((....))))).))))...))).))))(((((.........))))).... ( -28.20) >DroSec_CAF1 34112 120 + 1 GAUAUUUCCCCUAGAUGUAGAGGCGCAGCGGAAGUAUAACGUAUCGUUCUACAGGACACACAUGAACAGGGUCAUGACCCUAUUCACCAUUCUCUGCAUGACCUACACCUCGUCAUUUAG ..........(((((((..((((.((((.(((((((.((((...)))).)))..........((((.(((((....))))).))))...)))))))).((.....))))))..))))))) ( -32.60) >DroSim_CAF1 35683 120 + 1 GAUAUUUCCCCUAGAUGUAGAGGCGCAGCGGAAGUAUAACGUAUCGUUCUACAGGACACACAUGAACAGGGUCAUGACCCUAUUCACUAUCCUCUGCAUGACCUACACCUCGUCAUUUAG ..........(((((((..((((.((((.((((((..........((((....))))......(((.(((((....))))).)))))).))).)))).((.....))))))..))))))) ( -35.40) >DroEre_CAF1 32261 120 + 1 GAUAUUUCCUCUAGACGUUGAGAUGCAGCGGAAGUAUAACGUAUCGUUCUACAGGAAACACAUGAACAGGGUCAUGACCCUAUUUACCAUCCUCUGCAUGACCUACACCUCGUCAUUUAG .............((((.((((((((((.(((.(((.((((...)))).))).(....)...((((.(((((....))))).))))...))).))))))...)).))...))))...... ( -30.90) >DroYak_CAF1 47314 120 + 1 GAUCUUUCCCCUAGAUGUGGAGGCGCAGCGAAAGUAUAACGUAUCGUUCUACAGGACACACAUGAAUAGGGUCAUGACCCUAUUCACCAUCCUCUGCAUGACGUACACCUCGUCAUUUAG ..........(((((((..((((....((....))..........((((....)))).....((((((((((....))))))))))....))))..))(((((.......)))))))))) ( -37.00) >consensus GAUAUUUCCCCUAGAUGUAGAGGCGCAGCGGAAGUAUAACGUAUCGUUCUACAGGACACACAUGAACAGGGUCAUGACCCUAUUCACCAUCCUCUGCAUGACCUACACCUCGUCAUUUAG ..........(((((((..((((.((((.(((.(((.((((...)))).)))..........((((.(((((....))))).))))...))).)))).((.....))))))..))))))) (-28.26 = -28.82 + 0.56)


| Location | 16,330,875 – 16,330,995 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.42 |
| Mean single sequence MFE | -39.20 |
| Consensus MFE | -35.54 |
| Energy contribution | -36.18 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.50 |
| SVM RNA-class probability | 0.999314 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16330875 120 - 27905053 CUAAAUGACGAGGUGUAGGUCAUGCAGAGGAUGGUGAAUAGGGUCAUGACCCUGUUCAUGUGUUUCCGGUAAAACGAUACGUUAUACUUCCGCUGCGCUUCUAAAUCUAAAGGAAAUAUC ..........((((.((((...(((((.(((..(((((((((((....)))))))))))........((((.((((...)))).))))))).)))))..)))).))))............ ( -36.20) >DroSec_CAF1 34112 120 - 1 CUAAAUGACGAGGUGUAGGUCAUGCAGAGAAUGGUGAAUAGGGUCAUGACCCUGUUCAUGUGUGUCCUGUAGAACGAUACGUUAUACUUCCGCUGCGCCUCUACAUCUAGGGGAAAUAUC (((.(((..(((((((((....(((((.((...(((((((((((....))))))))))).....))))))).((((...)))).........)))))))))..))).))).......... ( -41.10) >DroSim_CAF1 35683 120 - 1 CUAAAUGACGAGGUGUAGGUCAUGCAGAGGAUAGUGAAUAGGGUCAUGACCCUGUUCAUGUGUGUCCUGUAGAACGAUACGUUAUACUUCCGCUGCGCCUCUACAUCUAGGGGAAAUAUC (((.(((..(((((((((.........(((((((((((((((((....)))))))))))...))))))(((.((((...)))).))).....)))))))))..))).))).......... ( -43.50) >DroEre_CAF1 32261 120 - 1 CUAAAUGACGAGGUGUAGGUCAUGCAGAGGAUGGUAAAUAGGGUCAUGACCCUGUUCAUGUGUUUCCUGUAGAACGAUACGUUAUACUUCCGCUGCAUCUCAACGUCUAGAGGAAAUAUC (((.(((..(((((((((....(((((..((((((.((((((((....)))))))).)).))))..))))).((((...)))).........)))))))))..))).))).......... ( -35.30) >DroYak_CAF1 47314 120 - 1 CUAAAUGACGAGGUGUACGUCAUGCAGAGGAUGGUGAAUAGGGUCAUGACCCUAUUCAUGUGUGUCCUGUAGAACGAUACGUUAUACUUUCGCUGCGCCUCCACAUCUAGGGGAAAGAUC (((.(((..((((((((((........(((((((((((((((((....)))))))))))...))))))(((.((((...)))).)))...)).))))))))..))).))).......... ( -39.90) >consensus CUAAAUGACGAGGUGUAGGUCAUGCAGAGGAUGGUGAAUAGGGUCAUGACCCUGUUCAUGUGUGUCCUGUAGAACGAUACGUUAUACUUCCGCUGCGCCUCUACAUCUAGGGGAAAUAUC (((.(((..(((((((((.........(((((((((((((((((....)))))))))))...))))))(((.((((...)))).))).....)))))))))..))).))).......... (-35.54 = -36.18 + 0.64)



| Location | 16,330,915 – 16,331,035 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.58 |
| Mean single sequence MFE | -33.52 |
| Consensus MFE | -30.50 |
| Energy contribution | -30.34 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.958998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16330915 120 - 27905053 AAGGUAAUACUUUAUGGUCGACUUGAUGGCUGGAUAAAAGCUAAAUGACGAGGUGUAGGUCAUGCAGAGGAUGGUGAAUAGGGUCAUGACCCUGUUCAUGUGUUUCCGGUAAAACGAUAC ..((.(((((..((((..(..((((.(((((.......))))).....)))).....)..)))).........(((((((((((....)))))))))))))))).))............. ( -33.70) >DroSec_CAF1 34152 120 - 1 AAGGUAAUACUUGAUGGUCGACUUGAUGGCUGGAUAAAAGCUAAAUGACGAGGUGUAGGUCAUGCAGAGAAUGGUGAAUAGGGUCAUGACCCUGUUCAUGUGUGUCCUGUAGAACGAUAC (((......)))....((((.((((.(((((.......))))).....))))...((((.((((((........((((((((((....)))))))))))))))).)))).....)))).. ( -33.80) >DroSim_CAF1 35723 120 - 1 AAGGUAAUACUUAAUGGUCGACUUGAUGGCUGGAUAAAAGCUAAAUGACGAGGUGUAGGUCAUGCAGAGGAUAGUGAAUAGGGUCAUGACCCUGUUCAUGUGUGUCCUGUAGAACGAUAC ...((..(((...(((..(..((((.(((((.......))))).....)))).....)..)))....(((((((((((((((((....)))))))))))...)))))))))..))..... ( -35.30) >DroEre_CAF1 32301 120 - 1 GAGGUAGUACUUAAUGGUCGACUUGAUGGCUGGAUAAAAGCUAAAUGACGAGGUGUAGGUCAUGCAGAGGAUGGUAAAUAGGGUCAUGACCCUGUUCAUGUGUUUCCUGUAGAACGAUAC (((......)))....((((......(((((.......))))).(((((.........)))))((((..((((((.((((((((....)))))))).)).))))..))))....)))).. ( -28.90) >DroYak_CAF1 47354 120 - 1 GAGGUAGUACUUAAUGGUCGACUUGAUGGCUGGAUAAAAGCUAAAUGACGAGGUGUACGUCAUGCAGAGGAUGGUGAAUAGGGUCAUGACCCUAUUCAUGUGUGUCCUGUAGAACGAUAC (((......)))....((((......(((((.......))))).((((((.......))))))..(.(((((((((((((((((....)))))))))))...)))))).)....)))).. ( -35.90) >consensus AAGGUAAUACUUAAUGGUCGACUUGAUGGCUGGAUAAAAGCUAAAUGACGAGGUGUAGGUCAUGCAGAGGAUGGUGAAUAGGGUCAUGACCCUGUUCAUGUGUGUCCUGUAGAACGAUAC (((......)))....((((......(((((.......))))).(((((.........)))))((((.((...(((((((((((....))))))))))).....))))))....)))).. (-30.50 = -30.34 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:17:02 2006