
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,329,527 – 16,329,658 |
| Length | 131 |
| Max. P | 0.867972 |

| Location | 16,329,527 – 16,329,618 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 79.14 |
| Mean single sequence MFE | -27.88 |
| Consensus MFE | -12.00 |
| Energy contribution | -13.37 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.48 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.810570 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16329527 91 + 27905053 UUGCUCGCCAACUCGGUGCACACGAACUCCUCAUCAGCUUAUCAGCUGCCCACGUUGCGUAUACGUUGCGUAUACGCCGCGUAUCGUGACU .(((..(((.....))))))(((((.........(((((....)))))...((((.(((((((((...))))))))).)))).)))))... ( -32.60) >DroSec_CAF1 32745 91 + 1 UUGCUCGCCAACUCGGUGCACACGACCUCCUCAUCAGCUUAUCAGCUGCCCACGUUGCGUAUACGUUGCGUAUACGCCGCGUAUCGUGACU .(((..(((.....))))))(((((.........(((((....)))))...((((.(((((((((...))))))))).)))).)))))... ( -32.30) >DroSim_CAF1 34384 91 + 1 UUGCUCGCCAACUCGGCGCACACGACCUCCUCAUCAGCUUAUCAGCUGCCCACGUUGCGUAUACGUUGCGUAUACGCCGCGUAUCGUGACU .....((((.....))))..(((((.........(((((....)))))...((((.(((((((((...))))))))).)))).)))))... ( -33.70) >DroEre_CAF1 30977 80 + 1 UUGCUCGCCAACUCGCUGCACACGACCUUCUCAUCAGCUUAUCAGCUGCCCACGUUGC-----------GUAUACGCCGUGUAUCGUGACU .(((..((......)).)))(((((.........(((((....)))))..((((..((-----------(....)))))))..)))))... ( -20.90) >DroYak_CAF1 45877 80 + 1 UUGCUCGCCAACUCGCUGCACACGGCCUUCUCAUCAGCUCAUCAGCUGCCCACGUUGC-----------GUAUACGCCGCGUAUCGUGACU .(((..((......)).)))(((((.........(((((....)))))...((((.((-----------(....))).)))).)))))... ( -21.70) >DroAna_CAF1 26878 80 + 1 UCGCUUGCCAACUCGAUGCACACGUCUUUUCGAUUGGCUUAUCAAUCG-----------AAUGCGUUGCGUAUACGCCGUGCGGUAUGAGU ..........(((((.(((((((((...(((((((((....)))))))-----------)).)))).(((....))).)))))...))))) ( -26.10) >consensus UUGCUCGCCAACUCGGUGCACACGACCUCCUCAUCAGCUUAUCAGCUGCCCACGUUGC_UAUACGUUGCGUAUACGCCGCGUAUCGUGACU ..(((((......))).)).(((((.........(((((....))))).............(((((.(((....))).))))))))))... (-12.00 = -13.37 + 1.36)


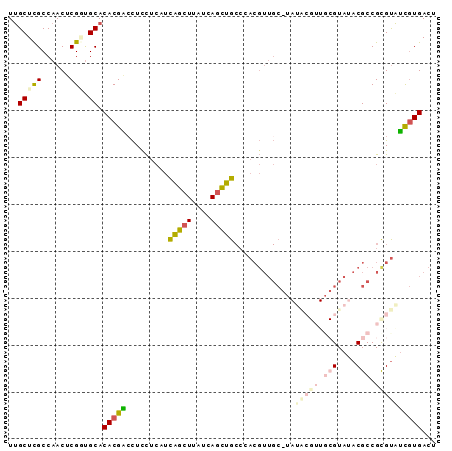
| Location | 16,329,527 – 16,329,618 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 79.14 |
| Mean single sequence MFE | -29.62 |
| Consensus MFE | -15.80 |
| Energy contribution | -16.37 |
| Covariance contribution | 0.57 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.555094 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16329527 91 - 27905053 AGUCACGAUACGCGGCGUAUACGCAACGUAUACGCAACGUGGGCAGCUGAUAAGCUGAUGAGGAGUUCGUGUGCACCGAGUUGGCGAGCAA ..(((...((((..(((((((((...)))))))))..))))..(((((....))))).)))...((((((..((.....))..)))))).. ( -33.40) >DroSec_CAF1 32745 91 - 1 AGUCACGAUACGCGGCGUAUACGCAACGUAUACGCAACGUGGGCAGCUGAUAAGCUGAUGAGGAGGUCGUGUGCACCGAGUUGGCGAGCAA ...(((((((((..(((((((((...)))))))))..)))...(((((....)))))........))))))(((.((.....))...))). ( -32.40) >DroSim_CAF1 34384 91 - 1 AGUCACGAUACGCGGCGUAUACGCAACGUAUACGCAACGUGGGCAGCUGAUAAGCUGAUGAGGAGGUCGUGUGCGCCGAGUUGGCGAGCAA ...(((((((((..(((((((((...)))))))))..)))...(((((....)))))........))))))..((((.....))))..... ( -36.20) >DroEre_CAF1 30977 80 - 1 AGUCACGAUACACGGCGUAUAC-----------GCAACGUGGGCAGCUGAUAAGCUGAUGAGAAGGUCGUGUGCAGCGAGUUGGCGAGCAA ...((((((.(((((((....)-----------))..))))..(((((....)))))........))))))(((.((......))..))). ( -26.40) >DroYak_CAF1 45877 80 - 1 AGUCACGAUACGCGGCGUAUAC-----------GCAACGUGGGCAGCUGAUGAGCUGAUGAGAAGGCCGUGUGCAGCGAGUUGGCGAGCAA .((((((.((((((((...(((-----------(...))))..(((((....)))))........))))))))...))...))))...... ( -24.30) >DroAna_CAF1 26878 80 - 1 ACUCAUACCGCACGGCGUAUACGCAACGCAUU-----------CGAUUGAUAAGCCAAUCGAAAAGACGUGUGCAUCGAGUUGGCAAGCGA ((((....((((((((((.......)))).((-----------((((((......))))))))....))))))....)))).......... ( -25.00) >consensus AGUCACGAUACGCGGCGUAUACGCAACGUAUA_GCAACGUGGGCAGCUGAUAAGCUGAUGAGAAGGUCGUGUGCACCGAGUUGGCGAGCAA .((((....((.(((.(((((((..........((.......))((((....))))...........))))))).))).))))))...... (-15.80 = -16.37 + 0.57)


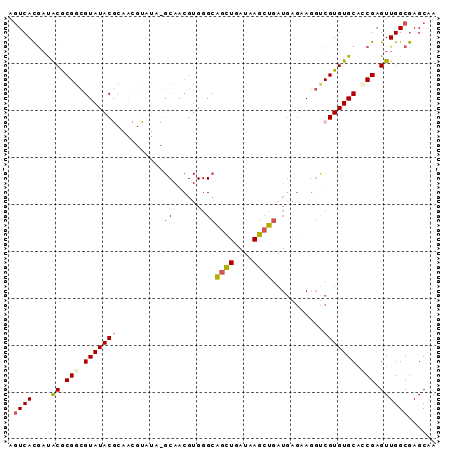
| Location | 16,329,547 – 16,329,658 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 90.17 |
| Mean single sequence MFE | -26.78 |
| Consensus MFE | -20.14 |
| Energy contribution | -22.22 |
| Covariance contribution | 2.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.632043 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16329547 111 + 27905053 CACGAACUCCUCAUCAGCUUAUCAGCUGCCCACGUUGCGUAUACGUUGCGUAUACGCCGCGUAUCGUGACUCAGCCCACAGCCCAGAAGCACACAAUAAAAAGCAUGAAAA (((((.........(((((....)))))...((((.(((((((((...))))))))).)))).)))))............((......))..................... ( -28.90) >DroSec_CAF1 32765 111 + 1 CACGACCUCCUCAUCAGCUUAUCAGCUGCCCACGUUGCGUAUACGUUGCGUAUACGCCGCGUAUCGUGACUCAGCCAACAGCCCAGAAGCACACAAUAAAAAGCAUGAAAA .............(((((((....((((.....((..((.((((((.(((....))).))))))))..)).)))).....((......))..........)))).)))... ( -28.80) >DroSim_CAF1 34404 111 + 1 CACGACCUCCUCAUCAGCUUAUCAGCUGCCCACGUUGCGUAUACGUUGCGUAUACGCCGCGUAUCGUGACUCAGCCAACAGCCCAGAAGCACACAAUAAAAAGCAUGAAAA .............(((((((....((((.....((..((.((((((.(((....))).))))))))..)).)))).....((......))..........)))).)))... ( -28.80) >DroEre_CAF1 30997 100 + 1 CACGACCUUCUCAUCAGCUUAUCAGCUGCCCACGUUGC-----------GUAUACGCCGUGUAUCGUGACUCAGCCGACAGCCCAGAAGCACACAAUAAAAAGCAUGAGAC ........((((((..((((.((.((((.....((..(-----------(.((((.....))))))..)).)))).))..((......))..........)))))))))). ( -23.30) >DroYak_CAF1 45897 100 + 1 CACGGCCUUCUCAUCAGCUCAUCAGCUGCCCACGUUGC-----------GUAUACGCCGCGUAUCGUGACUCAGCCGACAGCCCAGAAGCACACAAUAAAAAGCGUGAAAA ..((((....(((((((((....)))))...((((.((-----------(....))).))))...))))....))))............(((.(........).))).... ( -24.10) >consensus CACGACCUCCUCAUCAGCUUAUCAGCUGCCCACGUUGCGUAUACGUUGCGUAUACGCCGCGUAUCGUGACUCAGCCAACAGCCCAGAAGCACACAAUAAAAAGCAUGAAAA (((((.........(((((....)))))...((((.((((((((.....)))))))).)))).)))))............((......))..................... (-20.14 = -22.22 + 2.08)

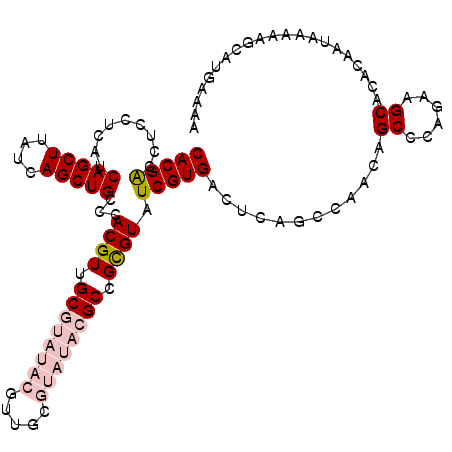

| Location | 16,329,547 – 16,329,658 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 90.17 |
| Mean single sequence MFE | -37.24 |
| Consensus MFE | -32.10 |
| Energy contribution | -33.82 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.867972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16329547 111 - 27905053 UUUUCAUGCUUUUUAUUGUGUGCUUCUGGGCUGUGGGCUGAGUCACGAUACGCGGCGUAUACGCAACGUAUACGCAACGUGGGCAGCUGAUAAGCUGAUGAGGAGUUCGUG .......((((((((((....((((.(.((((((.(((...)))....((((..(((((((((...)))))))))..)))).)))))).).)))).))))))))))..... ( -39.00) >DroSec_CAF1 32765 111 - 1 UUUUCAUGCUUUUUAUUGUGUGCUUCUGGGCUGUUGGCUGAGUCACGAUACGCGGCGUAUACGCAACGUAUACGCAACGUGGGCAGCUGAUAAGCUGAUGAGGAGGUCGUG ....(((((((((((((....((((.(.((((((((((...)))....((((..(((((((((...)))))))))..))))))))))).).)))).)))))))))..)))) ( -39.40) >DroSim_CAF1 34404 111 - 1 UUUUCAUGCUUUUUAUUGUGUGCUUCUGGGCUGUUGGCUGAGUCACGAUACGCGGCGUAUACGCAACGUAUACGCAACGUGGGCAGCUGAUAAGCUGAUGAGGAGGUCGUG ....(((((((((((((....((((.(.((((((((((...)))....((((..(((((((((...)))))))))..))))))))))).).)))).)))))))))..)))) ( -39.40) >DroEre_CAF1 30997 100 - 1 GUCUCAUGCUUUUUAUUGUGUGCUUCUGGGCUGUCGGCUGAGUCACGAUACACGGCGUAUAC-----------GCAACGUGGGCAGCUGAUAAGCUGAUGAGAAGGUCGUG ....(((((((((((((....((((.(.((((((((((...)))......(((((((....)-----------))..))))))))))).).)))).)))))))))..)))) ( -32.50) >DroYak_CAF1 45897 100 - 1 UUUUCACGCUUUUUAUUGUGUGCUUCUGGGCUGUCGGCUGAGUCACGAUACGCGGCGUAUAC-----------GCAACGUGGGCAGCUGAUGAGCUGAUGAGAAGGCCGUG ....(((((((((((((....((((.(.((((((((((...)))....((((..(((....)-----------))..))))))))))).).)))).)))))))))..)))) ( -35.90) >consensus UUUUCAUGCUUUUUAUUGUGUGCUUCUGGGCUGUCGGCUGAGUCACGAUACGCGGCGUAUACGCAACGUAUACGCAACGUGGGCAGCUGAUAAGCUGAUGAGGAGGUCGUG ....(((((((((((((....((((.(.((((((((((...)))......((((((((((((.....))))))))..))))))))))).).)))).)))))))))..)))) (-32.10 = -33.82 + 1.72)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:16:57 2006