| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,307,491 – 16,307,583 |
| Length | 92 |
| Max. P | 0.995394 |

| Location | 16,307,491 – 16,307,583 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 91.64 |
| Mean single sequence MFE | -20.49 |
| Consensus MFE | -17.76 |
| Energy contribution | -17.88 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.87 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
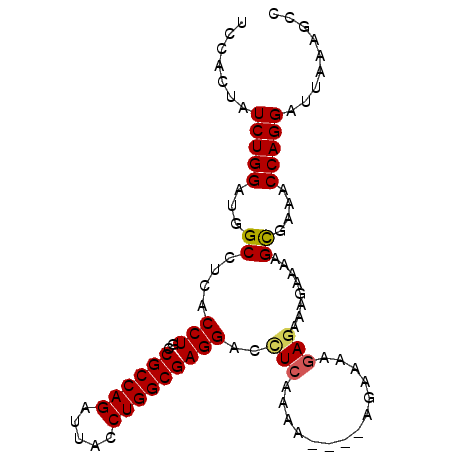
>3R_DroMel_CAF1 16307491 92 + 27905053 UCCACUAUCUGGAUAGCCUCACCUGCCGCCAGAUUACCUGGCGAGGACCUAAAAA----AAAAAAGAGAAAGAAAAGCGAAACCAGGAUUAAAGCC .......(((((...((....(((..((((((.....)))))))))..((.....----.....))..........))....)))))......... ( -18.20) >DroSec_CAF1 5097 96 + 1 UCCACUAUCUGGAUGGCCUCACCUGCCGCCAGAUUACCUGGCGAGGACCUCAAAAAAAAAGAAAAGAGAAAGAAAAGCGAAACCAGGAUUAAAGCC .......(((((.(.((....(((..((((((.....)))))))))..(((..............)))........)).)..)))))......... ( -20.64) >DroSim_CAF1 10256 95 + 1 UCCACUAUCUGGAUGGCCUCACCUGCCGCCAGAUUACCUGGCGAGGACCUCAAAA-AAAAGAAAAGAGAAAGAAAAGCGAAACCAGGAUUAAAGCC .......(((((.(.((....(((..((((((.....)))))))))..(((....-.........)))........)).)..)))))......... ( -20.72) >DroEre_CAF1 8233 89 + 1 UCUACUAUCUGGAUGGCCUCACCUGCCGCCAGAUUACCUGGCGAGGACUUCAAAA----A---AAGAGAAAGGAAAGCGAAACCAGGAUUAAAGCC ......((((((.((((.......))))))))))..(((((((....((((....----.---....).))).....))...)))))......... ( -20.10) >DroYak_CAF1 17095 92 + 1 UCCACUAUCUGGAUGGCCUCACCUGCCGCCAGAUUACCUGGCGAGGCUCUCAAAA----AGAAAUGAAAAAGGAAAGUGAAACCAGGAUUAAAGCC ((((.....))))(((..((((...(((((((.....)))))(((...)))....----............))...))))..)))........... ( -22.80) >consensus UCCACUAUCUGGAUGGCCUCACCUGCCGCCAGAUUACCUGGCGAGGACCUCAAAA____AGAAAAGAGAAAGAAAAGCGAAACCAGGAUUAAAGCC .......(((((...((....(((..((((((.....)))))))))..(((..............)))........))....)))))......... (-17.76 = -17.88 + 0.12)


| Location | 16,307,491 – 16,307,583 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 91.64 |
| Mean single sequence MFE | -27.81 |
| Consensus MFE | -25.36 |
| Energy contribution | -24.92 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.57 |
| SVM RNA-class probability | 0.995394 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
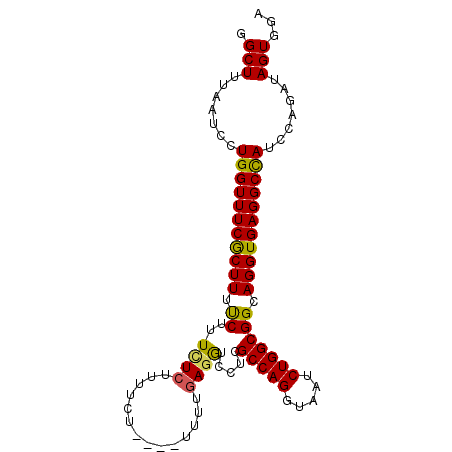
>3R_DroMel_CAF1 16307491 92 - 27905053 GGCUUUAAUCCUGGUUUCGCUUUUCUUUCUCUUUUUU----UUUUUAGGUCCUCGCCAGGUAAUCUGGCGGCAGGUGAGGCUAUCCAGAUAGUGGA (((((((..((((.....((((...............----.....))))..(((((((.....)))))))))))))))))).((((.....)))) ( -23.15) >DroSec_CAF1 5097 96 - 1 GGCUUUAAUCCUGGUUUCGCUUUUCUUUCUCUUUUCUUUUUUUUUGAGGUCCUCGCCAGGUAAUCUGGCGGCAGGUGAGGCCAUCCAGAUAGUGGA (((((((..((((..............((((..............))))...(((((((.....)))))))))))))))))).((((.....)))) ( -28.64) >DroSim_CAF1 10256 95 - 1 GGCUUUAAUCCUGGUUUCGCUUUUCUUUCUCUUUUCUUUU-UUUUGAGGUCCUCGCCAGGUAAUCUGGCGGCAGGUGAGGCCAUCCAGAUAGUGGA (((((((..((((..............((((.........-....))))...(((((((.....)))))))))))))))))).((((.....)))) ( -28.72) >DroEre_CAF1 8233 89 - 1 GGCUUUAAUCCUGGUUUCGCUUUCCUUUCUCUU---U----UUUUGAAGUCCUCGCCAGGUAAUCUGGCGGCAGGUGAGGCCAUCCAGAUAGUAGA .(((.......(((((((((((.((.....(((---(----....)))).....(((((.....))))))).))))))))))).......)))... ( -27.44) >DroYak_CAF1 17095 92 - 1 GGCUUUAAUCCUGGUUUCACUUUCCUUUUUCAUUUCU----UUUUGAGAGCCUCGCCAGGUAAUCUGGCGGCAGGUGAGGCCAUCCAGAUAGUGGA ...........(((((((((((.((.((((((.....----...))))))....(((((.....))))))).)))))))))))((((.....)))) ( -31.10) >consensus GGCUUUAAUCCUGGUUUCGCUUUUCUUUCUCUUUUCU____UUUUGAGGUCCUCGCCAGGUAAUCUGGCGGCAGGUGAGGCCAUCCAGAUAGUGGA .(((.......(((((((((((.((..((((..............)))).....(((((.....))))))).))))))))))).......)))... (-25.36 = -24.92 + -0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:16:48 2006