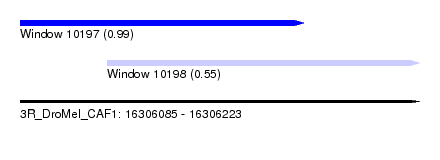
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,306,085 – 16,306,223 |
| Length | 138 |
| Max. P | 0.989857 |

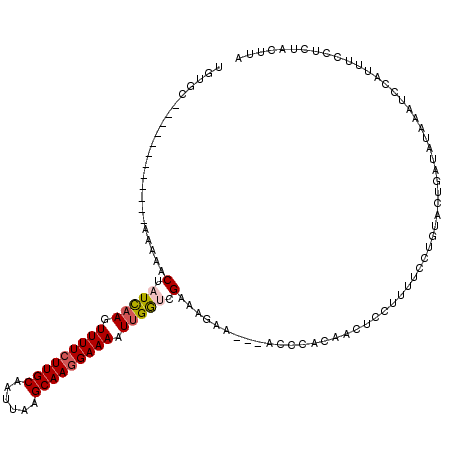
| Location | 16,306,085 – 16,306,183 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 75.61 |
| Mean single sequence MFE | -23.96 |
| Consensus MFE | -11.50 |
| Energy contribution | -11.90 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.48 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989857 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16306085 98 + 27905053 AACUGUAUCCAU---UUUCAGAAUGAAUUUUCCUGUUC------------AAAAACUAUCCAGUUUUCUUGCAAUUAAGCAAGGAAAAUUGGUCGAACUGCUUUUACCACAAC ............---...(((..(((((......))))------------)........(((((((((((((......)))).))))))))).....)))............. ( -18.50) >DroSec_CAF1 3721 95 + 1 AGCUGUAUUCAU---UUUCAGUAUGAAUUUUCCUGUGG------------AAAAACUAUCAAGUUUUCUUGCAAUUAAGCAAGGAAAAUUGGUCGAAAGCA---ACCCACAAC ......((((((---.......)))))).....(((((------------.......(((((.(((((((((......))))))))).)))))(....)..---..))))).. ( -22.50) >DroSim_CAF1 8859 95 + 1 AACUGUAUCCAU---UUUCAGUAUGAAUUUUCCUGUGC------------AAAAACUAUCAAGUUUUCUUGCAAUUAAGCAAGGAAAAUUGGUCGAAAGCC---ACCCACAAC .((((.......---...))))...(((((((((.(((------------(((((((....)))))..)))))........)))))))))((.(....)))---......... ( -20.30) >DroEre_CAF1 6873 108 + 1 AACGGUAUCCAUGUCGUUCAGAAUGAAUUUU-CUGUGCAGUGGGUGAUGGGAAAACUAUCAAGUUUUCUUGCAAUUAGGCAAGGAAAAUUGAACGAAAUAA---AU-AACCAC ...(((((((((...((.(((((......))-))).)).))))))(((((.....)))))...(((((((((......)))))))))..............---..-.))).. ( -30.10) >DroYak_CAF1 15691 109 + 1 UACUGUAUCCAUGGCUUUUGGCAUGAAUUUUCCUGUGCAGUGGCUGAUGGAAAAACAAUUAAGUUUUCUUGCAAUGAGGCAAAGAAAUUUGAACGACAGAA---AU-AAGCAC ..((((.(((((((((....(((((........)))))...)))).))))).......((((((((..((((......))))..))))))))...))))..---..-...... ( -28.40) >consensus AACUGUAUCCAU___UUUCAGAAUGAAUUUUCCUGUGC____________AAAAACUAUCAAGUUUUCUUGCAAUUAAGCAAGGAAAAUUGGUCGAAAGAA___ACCCACAAC .......................................................(.(((((.(((((((((......))))))))).))))).).................. (-11.50 = -11.90 + 0.40)

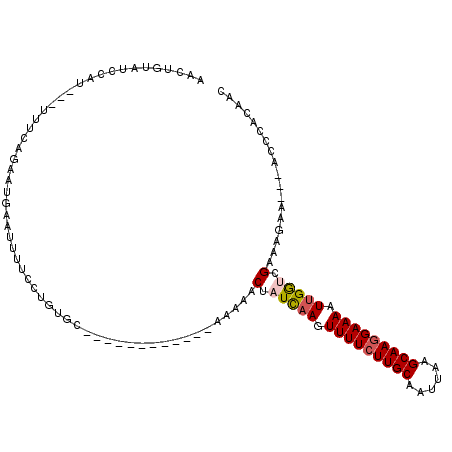
| Location | 16,306,115 – 16,306,223 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.42 |
| Mean single sequence MFE | -20.79 |
| Consensus MFE | -11.50 |
| Energy contribution | -11.90 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.553544 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16306115 108 + 27905053 UGUUC------------AAAAACUAUCCAGUUUUCUUGCAAUUAAGCAAGGAAAAUUGGUCGAACUGCUUUUACCACAACUGCUUUUCCUGUACUGAUAUAAAUUCAUUUUCUCUACUUA ...((------------(........(((((((((((((......)))).)))))))))..(((..((.............))..)))......)))....................... ( -16.72) >DroSec_CAF1 3751 105 + 1 UGUGG------------AAAAACUAUCAAGUUUUCUUGCAAUUAAGCAAGGAAAAUUGGUCGAAAGCA---ACCCACAACUCAUUUUCCUGUACUGAUAUAAAUCCAUUUCCUCUACUUA (((((------------.......(((((.(((((((((......))))))))).)))))(....)..---..))))).......................................... ( -20.10) >DroSim_CAF1 8889 105 + 1 UGUGC------------AAAAACUAUCAAGUUUUCUUGCAAUUAAGCAAGGAAAAUUGGUCGAAAGCC---ACCCACAACUCAUUUUCCUGUACUGAUAUAAAUCCAUUUCCUCUACUUA .((((------------(............(((((((((......)))))))))..(((.(....)))---).................))))).......................... ( -19.30) >DroEre_CAF1 6905 116 + 1 UGUGCAGUGGGUGAUGGGAAAACUAUCAAGUUUUCUUGCAAUUAGGCAAGGAAAAUUGAACGAAAUAA---AU-AACCACCUCCUCCCAUGUACUCAUAUAAAUCCUUUUCCUCAACUUA ......(((((((((((((......((((.(((((((((......))))))))).))))..((.....---..-.......)).)))))).)))))))...................... ( -26.84) >DroYak_CAF1 15724 116 + 1 UGUGCAGUGGCUGAUGGAAAAACAAUUAAGUUUUCUUGCAAUGAGGCAAAGAAAUUUGAACGACAGAA---AU-AAGCACUCCUUUGCCUGUAUUCAUAUAAAUUCCUUUCCUCAACUUA ..(((((.((((..((......))....))))...))))).(((((.((((.((((((((..((((..---..-..(((......))))))).))))....)))).)))))))))..... ( -21.00) >consensus UGUGC____________AAAAACUAUCAAGUUUUCUUGCAAUUAAGCAAGGAAAAUUGGUCGAAAGAA___ACCCACAACUCCUUUUCCUGUACUGAUAUAAAUCCAUUUCCUCUACUUA ......................(.(((((.(((((((((......))))))))).))))).).......................................................... (-11.50 = -11.90 + 0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:16:46 2006