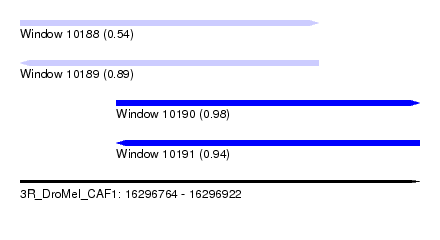
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,296,764 – 16,296,922 |
| Length | 158 |
| Max. P | 0.979187 |

| Location | 16,296,764 – 16,296,882 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -35.08 |
| Consensus MFE | -34.27 |
| Energy contribution | -33.50 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542296 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16296764 118 + 27905053 UCUAACAAACAUGAAUGGUUGCGUG--CUGUUCUAGGGUCAGAGUGUAAUUAACUUUGAAGCCCGCCCUCAUUUGCAGCCCAUCAAGGAGAUGAAGAGCAAAUUAGAGGUGCGGAAUAAG .........(..(((((((.....)--))))))...).(((((((.......)))))))...((((((((((((((....((((.....))))....))))))..)))).))))...... ( -34.30) >DroSec_CAF1 17428 120 + 1 UCCGGCAAACAUGAAUGGUUGCGUGCGCUGCUCUAGGGUCAAAGUGUAAUUAACUUUGAAGCCCGCCCUCAUUUGCAGCCCAUAAAGAAGAUGAAGAGCAAAUUAGAGGUGCGGAAUAAG (((.((((.((....)).))))..((((..(((((((((((((((.......))))))..))))(((.((((((..............)))))).).))....))))))))))))..... ( -34.44) >DroSim_CAF1 9587 120 + 1 UCCGGCAAACAUGAAUGGUUGCGUGCGCUGCUCUAGGGUCAAAGUGUAAUUAACUUUGAAGCCCGCCCUCAUUUGCAGCCCAUCAAGGAGAUGAAGAGCAAAUUAGAGGUGCGGAAUAAG (((((((..((((........))))...))))...((((((((((.......))))))..))))((((((((((((....((((.....))))....))))))..)))).)))))..... ( -36.50) >consensus UCCGGCAAACAUGAAUGGUUGCGUGCGCUGCUCUAGGGUCAAAGUGUAAUUAACUUUGAAGCCCGCCCUCAUUUGCAGCCCAUCAAGGAGAUGAAGAGCAAAUUAGAGGUGCGGAAUAAG (((((((..((((........))))...))))...((((((((((.......))))))..))))((((((((((((....((((.....))))....))))))..)))).)))))..... (-34.27 = -33.50 + -0.77)



| Location | 16,296,764 – 16,296,882 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -33.85 |
| Consensus MFE | -31.29 |
| Energy contribution | -31.73 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885979 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16296764 118 - 27905053 CUUAUUCCGCACCUCUAAUUUGCUCUUCAUCUCCUUGAUGGGCUGCAAAUGAGGGCGGGCUUCAAAGUUAAUUACACUCUGACCCUAGAACAG--CACGCAACCAUUCAUGUUUGUUAGA .....(((((.((((..((((((..((((((.....))))))..)))))))))))))))..................((((((....(((((.--..............))))))))))) ( -31.46) >DroSec_CAF1 17428 120 - 1 CUUAUUCCGCACCUCUAAUUUGCUCUUCAUCUUCUUUAUGGGCUGCAAAUGAGGGCGGGCUUCAAAGUUAAUUACACUUUGACCCUAGAGCAGCGCACGCAACCAUUCAUGUUUGCCGGA .....(((((.((((..((((((..(((((.......)))))..))))))))))))(((..(((((((.......)))))))))).......(.(((.(((........))).))))))) ( -32.70) >DroSim_CAF1 9587 120 - 1 CUUAUUCCGCACCUCUAAUUUGCUCUUCAUCUCCUUGAUGGGCUGCAAAUGAGGGCGGGCUUCAAAGUUAAUUACACUUUGACCCUAGAGCAGCGCACGCAACCAUUCAUGUUUGCCGGA .....(((((.((((..((((((..((((((.....))))))..))))))))))))(((..(((((((.......)))))))))).......(.(((.(((........))).))))))) ( -37.40) >consensus CUUAUUCCGCACCUCUAAUUUGCUCUUCAUCUCCUUGAUGGGCUGCAAAUGAGGGCGGGCUUCAAAGUUAAUUACACUUUGACCCUAGAGCAGCGCACGCAACCAUUCAUGUUUGCCGGA .....(((((.((((..((((((..((((((.....))))))..)))))))))))))))..(((((((.......)))))))................((((.((....)).)))).... (-31.29 = -31.73 + 0.45)

| Location | 16,296,802 – 16,296,922 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -30.90 |
| Consensus MFE | -30.47 |
| Energy contribution | -30.80 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16296802 120 + 27905053 AGAGUGUAAUUAACUUUGAAGCCCGCCCUCAUUUGCAGCCCAUCAAGGAGAUGAAGAGCAAAUUAGAGGUGCGGAAUAAGAAAUCCCAACGAAAGAAUGGAAAAGAAACAAAGACGCGUC ...((((......(((((....((((((((((((((....((((.....))))....))))))..)))).)))).........(((...(....)...))).......)))))))))... ( -32.10) >DroSec_CAF1 17468 120 + 1 AAAGUGUAAUUAACUUUGAAGCCCGCCCUCAUUUGCAGCCCAUAAAGAAGAUGAAGAGCAAAUUAGAGGUGCGGAAUAAGGAAUCCCGACGAAAGAAUGGAAAAGAAACAAAGACGCGUA ...((((......(((((....((((((((((((((....(((.......)))....))))))..)))).)))).........(((...(....)...))).......)))))))))... ( -28.50) >DroSim_CAF1 9627 120 + 1 AAAGUGUAAUUAACUUUGAAGCCCGCCCUCAUUUGCAGCCCAUCAAGGAGAUGAAGAGCAAAUUAGAGGUGCGGAAUAAGGAAUCCCGCCGAAAGAAUGGAAAAGAAACAAAGACGCGUC ...((((......(((((....((((((((((((((....((((.....))))....))))))..)))).)))).........(((...(....)...))).......)))))))))... ( -32.10) >consensus AAAGUGUAAUUAACUUUGAAGCCCGCCCUCAUUUGCAGCCCAUCAAGGAGAUGAAGAGCAAAUUAGAGGUGCGGAAUAAGGAAUCCCGACGAAAGAAUGGAAAAGAAACAAAGACGCGUC ...((((......(((((....((((((((((((((....((((.....))))....))))))..)))).)))).........(((...(....)...))).......)))))))))... (-30.47 = -30.80 + 0.33)



| Location | 16,296,802 – 16,296,922 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -30.50 |
| Consensus MFE | -28.70 |
| Energy contribution | -29.03 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16296802 120 - 27905053 GACGCGUCUUUGUUUCUUUUCCAUUCUUUCGUUGGGAUUUCUUAUUCCGCACCUCUAAUUUGCUCUUCAUCUCCUUGAUGGGCUGCAAAUGAGGGCGGGCUUCAAAGUUAAUUACACUCU ((.((.............(((((.........))))).........((((.((((..((((((..((((((.....))))))..)))))))))))))))).))................. ( -32.70) >DroSec_CAF1 17468 120 - 1 UACGCGUCUUUGUUUCUUUUCCAUUCUUUCGUCGGGAUUCCUUAUUCCGCACCUCUAAUUUGCUCUUCAUCUUCUUUAUGGGCUGCAAAUGAGGGCGGGCUUCAAAGUUAAUUACACUUU .......(((((.......(((...(....)...)))........(((((.((((..((((((..(((((.......)))))..)))))))))))))))...)))))............. ( -26.00) >DroSim_CAF1 9627 120 - 1 GACGCGUCUUUGUUUCUUUUCCAUUCUUUCGGCGGGAUUCCUUAUUCCGCACCUCUAAUUUGCUCUUCAUCUCCUUGAUGGGCUGCAAAUGAGGGCGGGCUUCAAAGUUAAUUACACUUU ..(((..........................(((((((.....))))))).((((..((((((..((((((.....))))))..)))))))))))))((((....))))........... ( -32.80) >consensus GACGCGUCUUUGUUUCUUUUCCAUUCUUUCGUCGGGAUUCCUUAUUCCGCACCUCUAAUUUGCUCUUCAUCUCCUUGAUGGGCUGCAAAUGAGGGCGGGCUUCAAAGUUAAUUACACUUU .......(((((......((((...........))))........(((((.((((..((((((..((((((.....))))))..)))))))))))))))...)))))............. (-28.70 = -29.03 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:16:39 2006