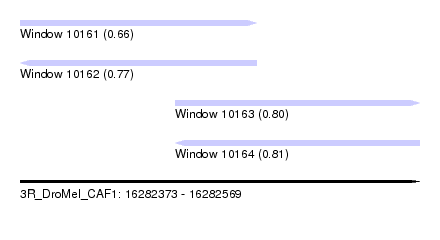
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,282,373 – 16,282,569 |
| Length | 196 |
| Max. P | 0.812828 |

| Location | 16,282,373 – 16,282,489 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.03 |
| Mean single sequence MFE | -26.89 |
| Consensus MFE | -21.46 |
| Energy contribution | -21.47 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.657314 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16282373 116 + 27905053 GCGGGCAUGGCACUCGGAAUAGCAAGCAAGUUGUUGUCCUUCCUGGUAGCAAAAGAAAAA----AAAAACAGGAAAAACUUACUUGCGAUUGCAAAGUAAAAGAAAAGCAGGCGACGAAG .((.((...((....(((.((((((.....)))))))))((((((...............----.....))))))....((((((((....)).)))))).......))..))..))... ( -24.35) >DroSec_CAF1 3105 120 + 1 GCGGGCAUUGCAUUCGGAAUAGCAAGCAAGUUGUUGUCCUUCCUGGUAGCAAAAAAAAAAAAGAAAAAACAGGAAAAACAUACUUGCAAUUGCAAAGUAAAAGAAAAGCAGGCGACGAAG .((.((.((((.(((......((((((((((...(((..((((((........................))))))..))).))))))..)))).........)))..))))))..))... ( -26.42) >DroYak_CAF1 4198 106 + 1 GCGGGGUCUGCAUUCGGAAUAGCAAGCAAGUUUUUGUCCUUCCUGCUGG--------------AAAAAACGGGGAAAACUUACUUGCGAUUGCAAAGUAAAAGAAAAGCAGGCGGCGAAG .((..((((((.(((......(((((.((((((((.((.((((....))--------------)).....)).)))))))).)))))..((((...))))..)))..))))))..))... ( -29.90) >consensus GCGGGCAUUGCAUUCGGAAUAGCAAGCAAGUUGUUGUCCUUCCUGGUAGCAAAA_AAAAA___AAAAAACAGGAAAAACUUACUUGCGAUUGCAAAGUAAAAGAAAAGCAGGCGACGAAG ((((...)))).((((.....((..((((((....((..((((((........................))))))..))..))))))..((((..............))))))..)))). (-21.46 = -21.47 + 0.01)

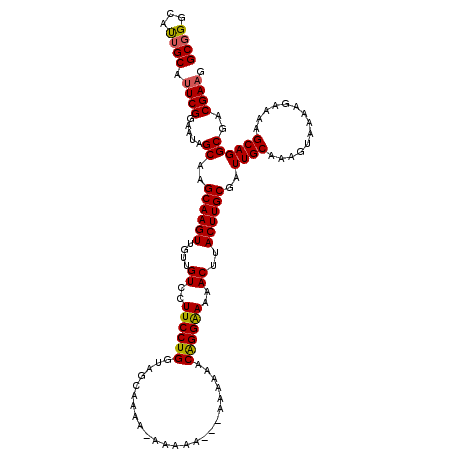

| Location | 16,282,373 – 16,282,489 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.03 |
| Mean single sequence MFE | -25.12 |
| Consensus MFE | -18.77 |
| Energy contribution | -19.22 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.769266 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16282373 116 - 27905053 CUUCGUCGCCUGCUUUUCUUUUACUUUGCAAUCGCAAGUAAGUUUUUCCUGUUUUU----UUUUUCUUUUGCUACCAGGAAGGACAACAACUUGCUUGCUAUUCCGAGUGCCAUGCCCGC .......((..((........(((((.(.(((.((((((((((((((((((.....----...............)))))))......))))))))))).)))).)))))....))..)) ( -23.75) >DroSec_CAF1 3105 120 - 1 CUUCGUCGCCUGCUUUUCUUUUACUUUGCAAUUGCAAGUAUGUUUUUCCUGUUUUUUCUUUUUUUUUUUUGCUACCAGGAAGGACAACAACUUGCUUGCUAUUCCGAAUGCAAUGCCCGC .......((.(((..(((.........((((..((((((.(((((((((((........................)))))))))))...))))))))))......))).)))..)).... ( -25.32) >DroYak_CAF1 4198 106 - 1 CUUCGCCGCCUGCUUUUCUUUUACUUUGCAAUCGCAAGUAAGUUUUCCCCGUUUUUU--------------CCAGCAGGAAGGACAAAAACUUGCUUGCUAUUCCGAAUGCAGACCCCGC .........((((...........((((.(((.(((((((((((((....(((((((--------------(.....)))))))).))))))))))))).))).)))).))))....... ( -26.30) >consensus CUUCGUCGCCUGCUUUUCUUUUACUUUGCAAUCGCAAGUAAGUUUUUCCUGUUUUUU___UUUUU_UUUUGCUACCAGGAAGGACAACAACUUGCUUGCUAUUCCGAAUGCAAUGCCCGC ...........((..(((.........((((..((((((..((((((((((........................))))))))))....))))))))))......))).))......... (-18.77 = -19.22 + 0.45)

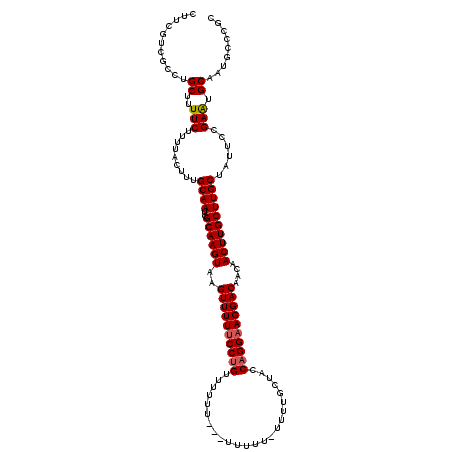
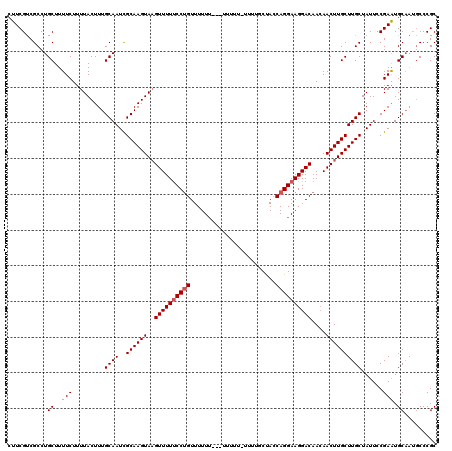
| Location | 16,282,449 – 16,282,569 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -33.29 |
| Consensus MFE | -31.75 |
| Energy contribution | -31.53 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799685 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16282449 120 + 27905053 UACUUGCGAUUGCAAAGUAAAAGAAAAGCAGGCGACGAAGAAGGAAGCCUCGAAUUGCUGCGGGAUUUCAUCUUCUUUCUUCUCGCACUUACUUUUCUUGUUUGCCACGUUUUUCAUGCU .....(((...(((((....((((((((.((((((.(((((((((((....(((((.(...).)))))...))))))))))))))).))..)))))))).)))))..))).......... ( -34.40) >DroSec_CAF1 3185 120 + 1 UACUUGCAAUUGCAAAGUAAAAGAAAAGCAGGCGACGAAGAAGGAAGCCUCGAAUUGCUGCGGGAUUUCAUCUUCUUUCUUCUCGCACUUACUUUUCUUGUUUGCCACGUUUUUCCUGCU (((((((....)).))))).((((((((.((((((.(((((((((((....(((((.(...).)))))...))))))))))))))).))..))))))))..................... ( -33.50) >DroYak_CAF1 4264 120 + 1 UACUUGCGAUUGCAAAGUAAAAGAAAAGCAGGCGGCGAAGAAGGAAGCCUCGAAUUGCUGCGGGAUUUUAUCUUCUUUCUUCUCGCACUUACUUUUCCUGUUUGCCACGUUUUUCCAGCU (((((((....)).)))))...(((((((.(..((((((..(((((((........))(((((((...............))))))).......))))).)))))).))))))))..... ( -31.96) >consensus UACUUGCGAUUGCAAAGUAAAAGAAAAGCAGGCGACGAAGAAGGAAGCCUCGAAUUGCUGCGGGAUUUCAUCUUCUUUCUUCUCGCACUUACUUUUCUUGUUUGCCACGUUUUUCCUGCU ...((((....)))).(((((.((((((.((((((.(((((((((((....(((((.(...).)))))...))))))))))))))).))..))))))...)))))............... (-31.75 = -31.53 + -0.22)


| Location | 16,282,449 – 16,282,569 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -33.14 |
| Consensus MFE | -30.85 |
| Energy contribution | -30.74 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812828 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16282449 120 - 27905053 AGCAUGAAAAACGUGGCAAACAAGAAAAGUAAGUGCGAGAAGAAAGAAGAUGAAAUCCCGCAGCAAUUCGAGGCUUCCUUCUUCGUCGCCUGCUUUUCUUUUACUUUGCAAUCGCAAGUA ..((((.....))))(((((.((((((((((.(.((((((((((.((((.......((((.(....).)).)))))).)))))).)))))))))))))))....)))))...(....).. ( -34.01) >DroSec_CAF1 3185 120 - 1 AGCAGGAAAAACGUGGCAAACAAGAAAAGUAAGUGCGAGAAGAAAGAAGAUGAAAUCCCGCAGCAAUUCGAGGCUUCCUUCUUCGUCGCCUGCUUUUCUUUUACUUUGCAAUUGCAAGUA .((((......)...(((((.((((((((((.(.((((((((((.((((.......((((.(....).)).)))))).)))))).)))))))))))))))....)))))...)))..... ( -34.01) >DroYak_CAF1 4264 120 - 1 AGCUGGAAAAACGUGGCAAACAGGAAAAGUAAGUGCGAGAAGAAAGAAGAUAAAAUCCCGCAGCAAUUCGAGGCUUCCUUCUUCGCCGCCUGCUUUUCUUUUACUUUGCAAUCGCAAGUA .((((........))))....((((((((((.(.(((.((((((.((((.......((((.(....).)).)))))).))))))..)))))))))))))).(((((.((....))))))) ( -31.41) >consensus AGCAGGAAAAACGUGGCAAACAAGAAAAGUAAGUGCGAGAAGAAAGAAGAUGAAAUCCCGCAGCAAUUCGAGGCUUCCUUCUUCGUCGCCUGCUUUUCUUUUACUUUGCAAUCGCAAGUA ...........((..(((((.((((((((((.(.((((((((((.((((.......((((.(....).)).)))))).)))))).)))))))))))))))....)))))...))...... (-30.85 = -30.74 + -0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:16:13 2006