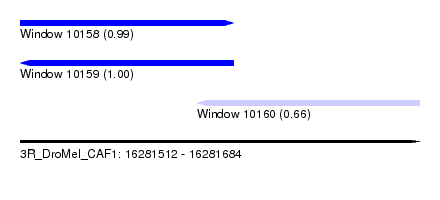
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,281,512 – 16,281,684 |
| Length | 172 |
| Max. P | 0.997700 |

| Location | 16,281,512 – 16,281,604 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.31 |
| Mean single sequence MFE | -35.20 |
| Consensus MFE | -27.51 |
| Energy contribution | -27.40 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986397 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16281512 92 + 27905053 UGCCAUGCCGGCAGGGCCAACAAGGACAACCAGUGCCACGACAAUGGUAAGAAAUGGAGUUGGCUGGCUGGAGGCG----------------------------UAGGAGGAGGACAGGA .(((...(((((..(((((((..((....))..(((((......))))).........))))))).))))).))).----------------------------................ ( -31.50) >DroSec_CAF1 2262 92 + 1 UGCCAUGCCGGCAGGGCCAACAAGGACAACCAGUGCCACGACAAUGGUAAGAAAUGGAGUUGGCUGGCUGGAGGCG----------------------------UAGGAGGAGGACAGGA .(((...(((((..(((((((..((....))..(((((......))))).........))))))).))))).))).----------------------------................ ( -31.50) >DroYak_CAF1 3270 120 + 1 UGCCAUGCCGGCAGCGCCAACAAGGACAACCACUGCCACGACAAUGGGGAGAAAUGGAGUUGGCUGGUUGGUGGCGUAGUAGGAGCUAGUUGGCGUAGAAGGCGUAGGAGGAGGAUUGGA ..(((((((..(.((((((((.....((((((..(((((..((.(.......).))..).)))))))))).((((.........)))))))))))).)..))))).))............ ( -42.60) >consensus UGCCAUGCCGGCAGGGCCAACAAGGACAACCAGUGCCACGACAAUGGUAAGAAAUGGAGUUGGCUGGCUGGAGGCG____________________________UAGGAGGAGGACAGGA .(((...(((((...((((((..((....))..(((((......))))).........))))))..))))).)))............................................. (-27.51 = -27.40 + -0.11)


| Location | 16,281,512 – 16,281,604 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.31 |
| Mean single sequence MFE | -27.28 |
| Consensus MFE | -25.29 |
| Energy contribution | -25.07 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.91 |
| SVM RNA-class probability | 0.997700 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16281512 92 - 27905053 UCCUGUCCUCCUCCUA----------------------------CGCCUCCAGCCAGCCAACUCCAUUUCUUACCAUUGUCGUGGCACUGGUUGUCCUUGUUGGCCCUGCCGGCAUGGCA ................----------------------------.(((.((.((..((((((..........((((.(((....))).)))).......))))))...)).))...))). ( -25.33) >DroSec_CAF1 2262 92 - 1 UCCUGUCCUCCUCCUA----------------------------CGCCUCCAGCCAGCCAACUCCAUUUCUUACCAUUGUCGUGGCACUGGUUGUCCUUGUUGGCCCUGCCGGCAUGGCA ................----------------------------.(((.((.((..((((((..........((((.(((....))).)))).......))))))...)).))...))). ( -25.33) >DroYak_CAF1 3270 120 - 1 UCCAAUCCUCCUCCUACGCCUUCUACGCCAACUAGCUCCUACUACGCCACCAACCAGCCAACUCCAUUUCUCCCCAUUGUCGUGGCAGUGGUUGUCCUUGUUGGCGCUGCCGGCAUGGCA .............(((.(((..(..(((((((.((..(..((((((((((...............................))))).))))).)..)).)))))))..)..))).))).. ( -31.18) >consensus UCCUGUCCUCCUCCUA____________________________CGCCUCCAGCCAGCCAACUCCAUUUCUUACCAUUGUCGUGGCACUGGUUGUCCUUGUUGGCCCUGCCGGCAUGGCA .............................................(((..((((((((((.(..((...........))..)))))..))))))....(((((((...))))))).))). (-25.29 = -25.07 + -0.22)



| Location | 16,281,588 – 16,281,684 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.55 |
| Mean single sequence MFE | -31.53 |
| Consensus MFE | -25.39 |
| Energy contribution | -25.50 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.658064 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16281588 96 - 27905053 CCAACUAUCUUGGCAGGCUGACUGCCUUCAGCGCUGGGCUCCGACUUGGUGGCAUCUUAUCCUGUUGCCGUCGUCGCGGUUCCUGUCCUCCUCCUA------------------------ ...........((((((((((......)))))...(((((.((((.(((..(((........)))..)))..)))).)))))))))).........------------------------ ( -31.20) >DroSec_CAF1 2338 96 - 1 CCAACUAUCUUGGCAGGCUGACUGCCUUCAGCGCUGGGCUCCGACUUGGUGGCAUCUUAUCCUGUCGCCGUCGUCGCGGUUCCUGUCCUCCUCCUA------------------------ ...........((((((((((......)))))...(((((.((((.((((((((........))))))))..)))).)))))))))).........------------------------ ( -32.70) >DroYak_CAF1 3350 117 - 1 CCAACUAUCUUGGCAGGCUGACUGCCUUCAGCGCUGGGCUCCGACUUAGUGGCAUCUUAUCCUGUCGCCG---UCGCGGUUCCAAUCCUCCUCCUACGCCUUCUACGCCAACUAGCUCCU ....(((..(((((((((.....))))...(((.((((...((((...((((((........)))))).)---))).((.......))....))))))).......))))).)))..... ( -30.70) >consensus CCAACUAUCUUGGCAGGCUGACUGCCUUCAGCGCUGGGCUCCGACUUGGUGGCAUCUUAUCCUGUCGCCGUCGUCGCGGUUCCUGUCCUCCUCCUA________________________ ((((.....))))((((((((......))))).)))((..(((...((((((((........))))))))......)))..))..................................... (-25.39 = -25.50 + 0.11)

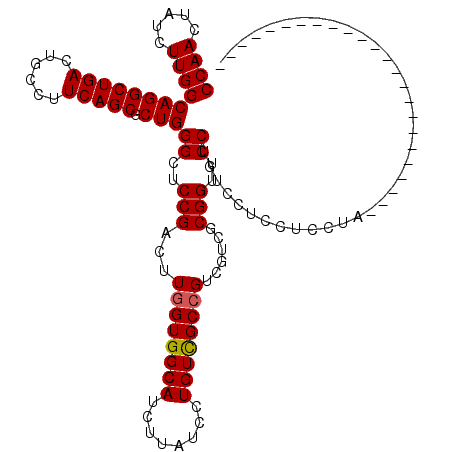

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:16:09 2006