
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,252,589 – 16,252,720 |
| Length | 131 |
| Max. P | 0.743943 |

| Location | 16,252,589 – 16,252,697 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 81.61 |
| Mean single sequence MFE | -28.78 |
| Consensus MFE | -21.17 |
| Energy contribution | -21.20 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.743943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
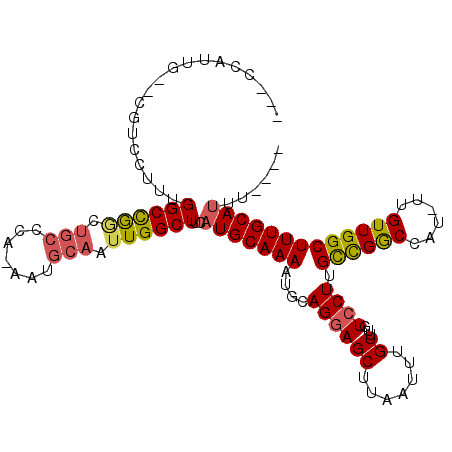
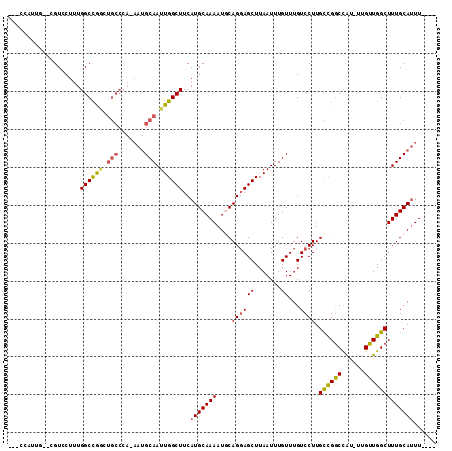
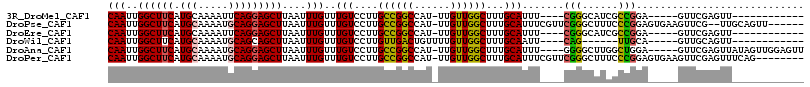
>3R_DroMel_CAF1 16252589 108 + 27905053 AGACCAUUGGAUGUGCUUUGGCCGGUUGCCCAAAAUGCAAUUGGCUUCAUGCAAAAUUCAGGAGCUUAAUUUGUUUGUCCUUGCCGGCCAU-UUGUUGGCUUUGCAUUU---- .(((..((((((.(((...((((((((((.......))))))))))....)))..))))))((((.......)))))))..(((.(((((.-....)))))..)))...---- ( -33.00) >DroPse_CAF1 64567 100 + 1 -----------CGUCCUUUGGCUGACUGCCCA-AAUGCAAUUGGCUUCAUGCAAAAUGCAGGAGCUUAAUUUGUUUGUCCUUGCCGGCCAU-UUGUUGGCUUUGCAUUUCGUU -----------........(((.....))).(-(((((((..((((((.(((.....)))))))))................((((((...-..)))))).)))))))).... ( -28.60) >DroEre_CAF1 54382 108 + 1 AGACCAUUGGAUGUGCUUUGGCCGGCUGCCCAAAAUGCAAUUGGCUUCAUGCAAAAUUCAGGAGCUUAAUUUGUUUGUCCUUGCCGGCCAU-UUGUUGGCUUUGCAUUU---- ........(((((((((.((((((((.(..(((...((((..((((((.((.......))))))))....)))))))..)..)))))))).-.....)))...))))))---- ( -31.00) >DroWil_CAF1 65681 94 + 1 UGAUUUUUG---GUCCUUUGGCUG------------GCAAUUGGCUUCAUGCAAAAUGCAGCAGCUUAAUUUGUUUGUCCUUGUUGACUGUUUUGUUGGCUUUGCAAUU---- ......(((---.......(((..------------((((..((((...(((.....)))..))))....))))..)))...((..((......))..))....)))..---- ( -19.40) >DroAna_CAF1 71180 105 + 1 ---CCACCGAACGUCCUUUGGCCAGUUGCCCAAAAUGCAAUUGGCUUCAUGCAAAAUGCAGGAGCUUAAUUUGUUUGUCCUUGCCGGCCAU-UUGUUGGCUUUGCAUUU---- ---................((((((((((.......))))))))))..(((((((..(((((..(...........)..)))))..((((.-....)))))))))))..---- ( -32.10) >DroPer_CAF1 69868 100 + 1 -----------CGUCCUUUGGCUGACUGCCCA-AAUGCAAUUGGCUUCAUGCAAAAUGCAGGAGCUUAAUUUGUUUGUCCUUGCCGGCCAU-UUGUUGGCUUUGCAUUUCGUU -----------........(((.....))).(-(((((((..((((((.(((.....)))))))))................((((((...-..)))))).)))))))).... ( -28.60) >consensus ___CCAUUG__CGUCCUUUGGCCGGCUGCCCA_AAUGCAAUUGGCUUCAUGCAAAAUGCAGGAGCUUAAUUUGUUUGUCCUUGCCGGCCAU_UUGUUGGCUUUGCAUUU____ ...................((((((.(((.......))).))))))..(((((((....((((((.......))...)))).((((((......)))))))))))))...... (-21.17 = -21.20 + 0.03)

| Location | 16,252,626 – 16,252,720 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 80.84 |
| Mean single sequence MFE | -28.12 |
| Consensus MFE | -18.91 |
| Energy contribution | -18.67 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519474 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
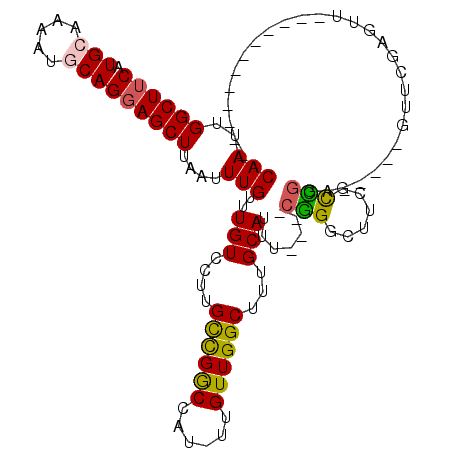
>3R_DroMel_CAF1 16252626 94 + 27905053 CAAUUGGCUUCAUGCAAAAUUCAGGAGCUUAAUUUGUUUGUCCUUGCCGGCCAU-UUGUUGGCUUUGCAUUU----CGGGCAUCGCCGGA-----GUUCGAGUU------------ ...(((((((((((((((....((((((.......))...)))).((((((...-..)))))))))))))..----..(((...))))))-----).))))...------------ ( -24.30) >DroPse_CAF1 64592 107 + 1 CAAUUGGCUUCAUGCAAAAUGCAGGAGCUUAAUUUGUUUGUCCUUGCCGGCCAU-UUGUUGGCUUUGCAUUUCGUUCGGGCUUUCCCGGAGUGAAGUUCG--UUGCAGUU------ ((((..((((((((((((..(((((..(...........)..)))))..((((.-....)))))))))......((((((....)))))))))))))..)--))).....------ ( -35.00) >DroEre_CAF1 54419 94 + 1 CAAUUGGCUUCAUGCAAAAUUCAGGAGCUUAAUUUGUUUGUCCUUGCCGGCCAU-UUGUUGGCUUUGCAUUU----CGGGCAUCGCCGGA-----GUUCGAGUU------------ ...(((((((((((((((....((((((.......))...)))).((((((...-..)))))))))))))..----..(((...))))))-----).))))...------------ ( -24.30) >DroWil_CAF1 65703 89 + 1 CAAUUGGCUUCAUGCAAAAUGCAGCAGCUUAAUUUGUUUGUCCUUGUUGACUGUUUUGUUGGCUUUGCAAUU----CAG------UUGCA-----GUUGCAGUU------------ .....((((...(((.....)))(((((((((((...((((....((..((......))..))...))))..----.))------))).)-----)))))))))------------ ( -20.70) >DroAna_CAF1 71214 106 + 1 CAAUUGGCUUCAUGCAAAAUGCAGGAGCUUAAUUUGUUUGUCCUUGCCGGCCAU-UUGUUGGCUUUGCAUUU----GGGGCUUGGCUGGA-----GUUCGAGUUAUAGUUGGAGUU (((((((((((.(((.....)))))))).(((((((....(((..(((((((..-.(((.......)))...----..))).)))).)))-----...)))))))))))))..... ( -32.20) >DroPer_CAF1 69893 107 + 1 CAAUUGGCUUCAUGCAAAAUGCAGGAGCUUAAUUUGUUUGUCCUUGCCGGCCAU-UUGUUGGCUUUGCAUUUCGUUCGGGCUUUCCCGGAGUGAAGUUCGAGUUUCAG-------- ...(((((((((((((((..(((((..(...........)..)))))..((((.-....)))))))))......((((((....)))))))))))).)))).......-------- ( -32.20) >consensus CAAUUGGCUUCAUGCAAAAUGCAGGAGCUUAAUUUGUUUGUCCUUGCCGGCCAU_UUGUUGGCUUUGCAUUU____CGGGCUUCGCCGGA_____GUUCGAGUU____________ (((..((((((.(((.....)))))))))....)))..(((....((((((......))))))...))).......(((......)))............................ (-18.91 = -18.67 + -0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:54 2006