| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,249,497 – 16,249,594 |
| Length | 97 |
| Max. P | 0.754040 |

| Location | 16,249,497 – 16,249,594 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 76.35 |
| Mean single sequence MFE | -17.29 |
| Consensus MFE | -12.32 |
| Energy contribution | -12.35 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.754040 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
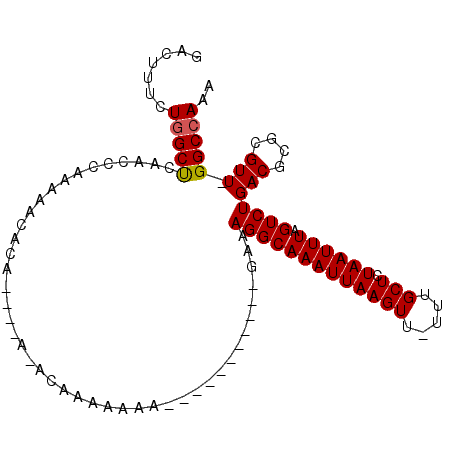
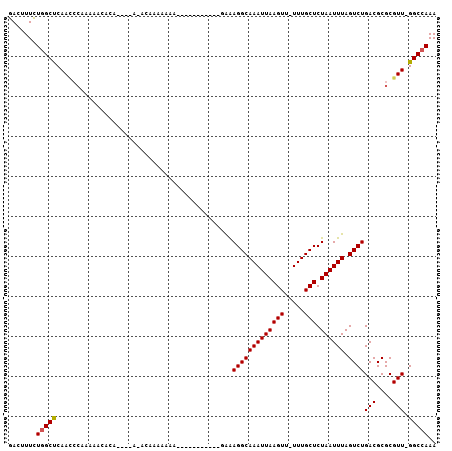
>3R_DroMel_CAF1 16249497 97 - 27905053 GACUUUCUGGCUCAACCCAAAAACACA----AAACAAAAAAAA----AAAAACGGAAGGCAAAUUAAGUU-UUUGCUCUAAUUUAGUCUGACGCGCGUU-GGCCAAA .......((((.((((.(.........----............----.....((..(((((((((((((.-...))).)))))).))))..)).).)))-))))).. ( -16.53) >DroPse_CAF1 61609 75 - 1 GACUUUCUGGCUCAAUCCGAAA-A------------------------------AAAGGCAAAUUAAGUUUUUUGCUCUAAUUUAGUCUGACGCGCGUU-GGCCAAA .......((((.((((.((...-.------------------------------..(((((((((((((.....))).)))))).))))....)).)))-))))).. ( -18.80) >DroSim_CAF1 73972 101 - 1 GACUUUCUGGCUCAACCCAAAAACACA----AAACAAAAAAAAAAAAAAAAAGGGAAGGCAAAUUAAGUU-UUUGCUCUAAUUUAGUCUGACGCGCGUU-GGCCAAA .......((((.((((((.........----.....................))).(((((((((((((.-...))).)))))).))))........))-))))).. ( -16.21) >DroEre_CAF1 51295 90 - 1 GACUUUCUGGCUCAACCCAAAAACACA----AGCCAAAAAAA-----------GGGAGGCAAAUUAAGUU-UUUGCUCUAAUUUAGUCUGACGCGCGUU-GGCAAAA .......(((......)))........----.(((((.....-----------(..(((((((((((((.-...))).)))))).))))..).....))-))).... ( -18.60) >DroAna_CAF1 62760 99 - 1 GACUUUUUGGCCCAA-----AUAUCCUCCUCCGACAAAAAAAA---AAACAGGCAAAGGCAAAUUAAGUUUUUUGCUCUAAUUUAGUCUGACGCGCGUUUGGCCAAA .....(((((((.((-----((....((....)).........---...((((((((((((((........))))))....))).)))))......))))))))))) ( -19.00) >DroPer_CAF1 66929 76 - 1 GACUUUCUGGCUCAAUCCAAAAAA------------------------------AAAGGCAAAUUAAGUUUUUUGCUCUAAUUUAGUCUGACGCGCGUU-GGCCAAA .......((((.((((........------------------------------..(((((((((((((.....))).)))))).)))).......)))-))))).. ( -14.63) >consensus GACUUUCUGGCUCAACCCAAAAACACA____A_ACAAAAAAA___________GAAAGGCAAAUUAAGUU_UUUGCUCUAAUUUAGUCUGACGCGCGUU_GGCCAAA .......(((((............................................(((((((((((((.....))).)))))).))))(((....))).))))).. (-12.32 = -12.35 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:52 2006