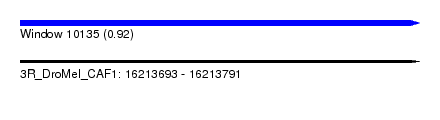
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,213,693 – 16,213,791 |
| Length | 98 |
| Max. P | 0.922595 |

| Location | 16,213,693 – 16,213,791 |
|---|---|
| Length | 98 |
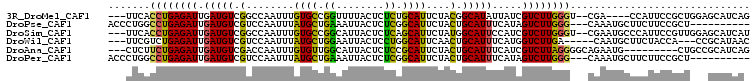
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 71.70 |
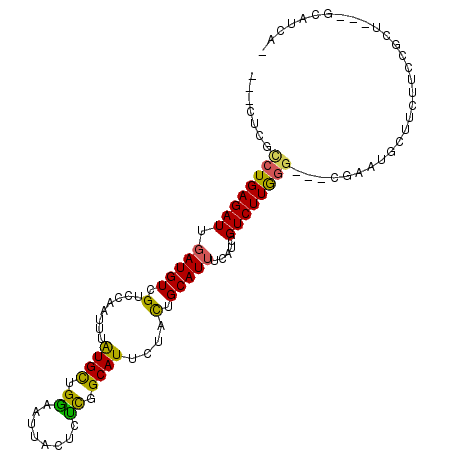
| Mean single sequence MFE | -26.92 |
| Consensus MFE | -11.61 |
| Energy contribution | -10.83 |
| Covariance contribution | -0.77 |
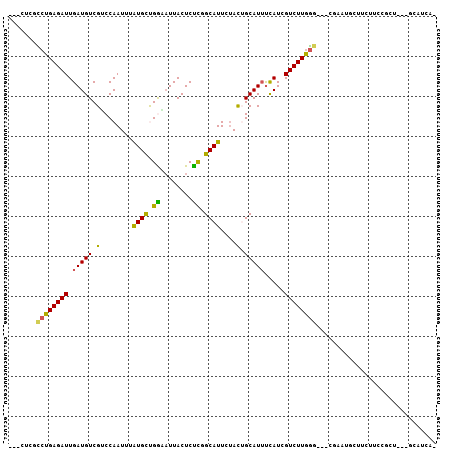
| Combinations/Pair | 1.40 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.43 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16213693 98 + 27905053 ---UUCACCUGAGAUUGAUGUCGGCCAAUUUGUGCCGGUUUUACUCUCUGCAUUCUACGGCAUAUUAUCGUCUUGGGU--CGA----CCAUUCCGCUGGAGCAUCAG ---.(((((..((((.((((...(((.....((((.((........)).)))).....)))....))))))))..)))--.))----(((......)))........ ( -25.10) >DroPse_CAF1 23322 94 + 1 ACCCUGGCCUGAGAUUGAUGUCGUCCAAUUUAUGCUGAAAUUACUCUCGGCAUUCUACUGCAUUUCAUAGUCUUGGG---CAAAUGCUUCUUCCGCU---------- ......(((..((((((.((..((.((....(((((((........))))))).....)).))..))))))))..))---)....((.......)).---------- ( -24.30) >DroSim_CAF1 12278 102 + 1 ---UUCACCUGAGAUUGAUGUCGGCCAAUUUGUGCCGGCAUUACUCUCAGCAUUCUAUGGCAUUCCAUCGUCUUGGGU--CGAAUGCCCAUUCCGUUGGAGCAUCAU ---.....((((((.(((((((((((.....).)))))))))).))))))......((((..(((((.((...(((((--.....)))))...)).)))))..)))) ( -39.90) >DroWil_CAF1 21255 96 + 1 ---UUCGUCUGAGAUUGAUGUCGUCCAAUUUAUGCUGGAAUUACUCCUGGCAUUCAACUGCAUUUCAUGGUCUUGA-----CAAUGCUUCUACCA---CCGCAUAAC ---...(((.(((((((((((.((.......((((((((.....))).)))))...)).)))))....))))))))-----).((((........---..))))... ( -19.10) >DroAna_CAF1 25909 95 + 1 ---CUCUUCUGAGAUUGAUGUCGACCAAUUUGUGUUGGCAUUACUCUCCGCAUUCUACUGCAUUUCAUCGUCUUAGGGGCAGAAUG---------CUGCCGCAUCAG ---.......((((.(((((((((((.....).)))))))))).)))).((..((((..((........))..))))(((((....---------)))))))..... ( -28.80) >DroPer_CAF1 28495 94 + 1 ACCCUGGCCUGAGAUUGAUGUCGUCCAAUUUAUGCUGAAAUUACUCUCGGCAUUCUACUGCAUUUCAUAGUCUUGGG---CAAAUGCUUCUUCCGCU---------- ......(((..((((((.((..((.((....(((((((........))))))).....)).))..))))))))..))---)....((.......)).---------- ( -24.30) >consensus ___CUCGCCUGAGAUUGAUGUCGUCCAAUUUAUGCUGGAAUUACUCUCGGCAUUCUACUGCAUUUCAUCGUCUUGGG___CGAAUGCUUCUUCCGCU___GCAUCA_ .......((((((((.(((((.(........((((.((........)).))))....).))))).....)))))))).............................. (-11.61 = -10.83 + -0.77)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:45 2006