
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,198,785 – 16,198,985 |
| Length | 200 |
| Max. P | 0.955458 |

| Location | 16,198,785 – 16,198,905 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -26.60 |
| Consensus MFE | -26.60 |
| Energy contribution | -26.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.53 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.955458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16198785 120 + 27905053 CUUUUGCUCUUUACUGGUCUGACUUUUUAGCAUAUUUCCCUUAGCAUAUUUUUAUUUGUAGCAUUUAUAAGUGAAUAAAUGCGCGCAUUUAAAUGCCACUUUCUGGCCCCGUCCAGCCAG .............(((((..(((...........................((((..(((.((((((((......))))))))..)))..)))).((((.....))))...)))..))))) ( -26.50) >DroSec_CAF1 27984 120 + 1 UUUUUGCUCUUUACUGGUCUGACUUUUUAGCAUAUUUCCCUUAGCAUAUUUUUAUUUGUAGCAUUUAUAAGUGAAUAAAUGCGCGCAUUUAAAUGCCACUUUCUGGCCCCGUCCAGCCAG .............(((((..(((...........................((((..(((.((((((((......))))))))..)))..)))).((((.....))))...)))..))))) ( -26.50) >DroEre_CAF1 28196 120 + 1 UUUUUGCUCUUUACUGGUCUGACUUUUUAGCAUAUUUCCCUUAGCAUAUUUUUAUUUGUAGCAUUUAUAAGUGAAUAAAUGCGCGCAUUUAAAUGCCACUUUCUGGCCCCGUCCAGCCAG .............(((((..(((...........................((((..(((.((((((((......))))))))..)))..)))).((((.....))))...)))..))))) ( -26.50) >DroYak_CAF1 30394 120 + 1 UUUUUGCACUUUACUGGUCUGACUUUUUAGCAUAUUUCCCUUAACAUAUUUUUAUUUGUAGCAUUUAUAAGUGAAUAAAUGCGCGCAUUUAAAUGCCACUUUCUGGCUCCGUCCAGCCAG .............(((((..(((...........................((((..(((.((((((((......))))))))..)))..)))).((((.....))))...)))..))))) ( -26.90) >consensus UUUUUGCUCUUUACUGGUCUGACUUUUUAGCAUAUUUCCCUUAGCAUAUUUUUAUUUGUAGCAUUUAUAAGUGAAUAAAUGCGCGCAUUUAAAUGCCACUUUCUGGCCCCGUCCAGCCAG .............(((((..(((...........................((((..(((.((((((((......))))))))..)))..)))).((((.....))))...)))..))))) (-26.60 = -26.60 + -0.00)



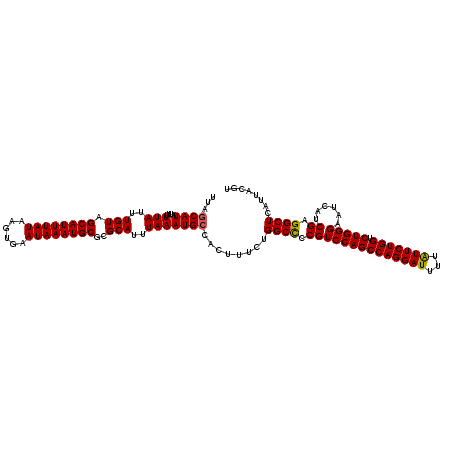
| Location | 16,198,825 – 16,198,945 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.75 |
| Mean single sequence MFE | -32.97 |
| Consensus MFE | -31.77 |
| Energy contribution | -31.65 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894392 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16198825 120 + 27905053 UUAGCAUAUUUUUAUUUGUAGCAUUUAUAAGUGAAUAAAUGCGCGCAUUUAAAUGCCACUUUCUGGCCCCGUCCAGCCAGGAUUUUAUUCUGGUCUGGGAAUCAUCGAGGCUCAUUACGU ...((((....(((..(((.((((((((......))))))))..)))..)))))))........((((.((((((((((((((...))))))).)))))......)).))))........ ( -33.30) >DroSec_CAF1 28024 120 + 1 UUAGCAUAUUUUUAUUUGUAGCAUUUAUAAGUGAAUAAAUGCGCGCAUUUAAAUGCCACUUUCUGGCCCCGUCCAGCCAGGAUUUUAUUCUGGUCUGGGAAUCAUCGAGGCUCAUUACGU ...((((....(((..(((.((((((((......))))))))..)))..)))))))........((((.((((((((((((((...))))))).)))))......)).))))........ ( -33.30) >DroEre_CAF1 28236 120 + 1 UUAGCAUAUUUUUAUUUGUAGCAUUUAUAAGUGAAUAAAUGCGCGCAUUUAAAUGCCACUUUCUGGCCCCGUCCAGCCAGGAUUUUGUUCUGGUCUGGGAAUCAUCGAGGCUCAUUACGU ...((((....(((..(((.((((((((......))))))))..)))..)))))))........((((.((((((((((((((...))))))).)))))......)).))))........ ( -33.60) >DroYak_CAF1 30434 120 + 1 UUAACAUAUUUUUAUUUGUAGCAUUUAUAAGUGAAUAAAUGCGCGCAUUUAAAUGCCACUUUCUGGCUCCGUCCAGCCAGGAUUUUAUUCUGGUCUGGGAAUCAUCGAGGCUCAUUACGU ..........((((..(((.((((((((......))))))))..)))..)))).((((.....))))..((((((((((((((...))))))).))))((..(.....)..))...))). ( -31.70) >consensus UUAGCAUAUUUUUAUUUGUAGCAUUUAUAAGUGAAUAAAUGCGCGCAUUUAAAUGCCACUUUCUGGCCCCGUCCAGCCAGGAUUUUAUUCUGGUCUGGGAAUCAUCGAGGCUCAUUACGU ...((((....(((..(((.((((((((......))))))))..)))..)))))))........((((.((((((((((((((...))))))).)))))......)).))))........ (-31.77 = -31.65 + -0.12)

| Location | 16,198,825 – 16,198,945 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.75 |
| Mean single sequence MFE | -31.77 |
| Consensus MFE | -31.35 |
| Energy contribution | -31.60 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952098 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16198825 120 - 27905053 ACGUAAUGAGCCUCGAUGAUUCCCAGACCAGAAUAAAAUCCUGGCUGGACGGGGCCAGAAAGUGGCAUUUAAAUGCGCGCAUUUAUUCACUUAUAAAUGCUACAAAUAAAAAUAUGCUAA .((((.((.(((((((....))((((.((((.........))))))))..)))))))....((((((((((((((....))))..........)))))))))).........)))).... ( -32.90) >DroSec_CAF1 28024 120 - 1 ACGUAAUGAGCCUCGAUGAUUCCCAGACCAGAAUAAAAUCCUGGCUGGACGGGGCCAGAAAGUGGCAUUUAAAUGCGCGCAUUUAUUCACUUAUAAAUGCUACAAAUAAAAAUAUGCUAA .((((.((.(((((((....))((((.((((.........))))))))..)))))))....((((((((((((((....))))..........)))))))))).........)))).... ( -32.90) >DroEre_CAF1 28236 120 - 1 ACGUAAUGAGCCUCGAUGAUUCCCAGACCAGAACAAAAUCCUGGCUGGACGGGGCCAGAAAGUGGCAUUUAAAUGCGCGCAUUUAUUCACUUAUAAAUGCUACAAAUAAAAAUAUGCUAA .((((.((.(((((((....))((((.((((.........))))))))..)))))))....((((((((((((((....))))..........)))))))))).........)))).... ( -32.90) >DroYak_CAF1 30434 120 - 1 ACGUAAUGAGCCUCGAUGAUUCCCAGACCAGAAUAAAAUCCUGGCUGGACGGAGCCAGAAAGUGGCAUUUAAAUGCGCGCAUUUAUUCACUUAUAAAUGCUACAAAUAAAAAUAUGUUAA (((((.((.((..(((....))((((.((((.........))))))))..)..))))....((((((((((((((....))))..........)))))))))).........)))))... ( -28.40) >consensus ACGUAAUGAGCCUCGAUGAUUCCCAGACCAGAAUAAAAUCCUGGCUGGACGGGGCCAGAAAGUGGCAUUUAAAUGCGCGCAUUUAUUCACUUAUAAAUGCUACAAAUAAAAAUAUGCUAA .((((.((.(((((((....))((((.((((.........))))))))..)))))))....((((((((((((((....))))..........)))))))))).........)))).... (-31.35 = -31.60 + 0.25)

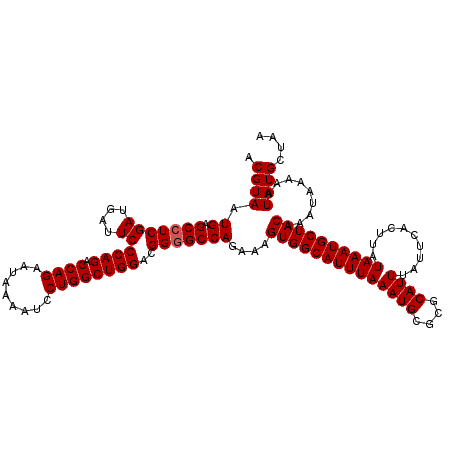

| Location | 16,198,865 – 16,198,985 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.17 |
| Mean single sequence MFE | -36.21 |
| Consensus MFE | -35.96 |
| Energy contribution | -36.21 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926703 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16198865 120 - 27905053 CGUGCAUAAAAUGCUGAGCACAAAAAAAUUUCCACUGCAUACGUAAUGAGCCUCGAUGAUUCCCAGACCAGAAUAAAAUCCUGGCUGGACGGGGCCAGAAAGUGGCAUUUAAAUGCGCGC (((((((.(((((((..(((...............)))..((....((.(((((((....))((((.((((.........))))))))..)))))))....)))))))))..))))))). ( -37.56) >DroSec_CAF1 28064 120 - 1 CGUGCAUAAAAUGCUGAGCACAAAAAAAUUUCCACUGCAUACGUAAUGAGCCUCGAUGAUUCCCAGACCAGAAUAAAAUCCUGGCUGGACGGGGCCAGAAAGUGGCAUUUAAAUGCGCGC (((((((.(((((((..(((...............)))..((....((.(((((((....))((((.((((.........))))))))..)))))))....)))))))))..))))))). ( -37.56) >DroEre_CAF1 28276 120 - 1 CGUGCAUAAAAUGCUGAGCACAAAAAAAUUUCCACUGCAUACGUAAUGAGCCUCGAUGAUUCCCAGACCAGAACAAAAUCCUGGCUGGACGGGGCCAGAAAGUGGCAUUUAAAUGCGCGC (((((((.(((((((..(((...............)))..((....((.(((((((....))((((.((((.........))))))))..)))))))....)))))))))..))))))). ( -37.56) >DroYak_CAF1 30474 120 - 1 CGUGCAUAAAAUGCUGAGCACAAAAAAAUUUCCACUGCAUACGUAAUGAGCCUCGAUGAUUCCCAGACCAGAAUAAAAUCCUGGCUGGACGGAGCCAGAAAGUGGCAUUUAAAUGCGCGC (((((((.(((((((..(((...............)))..((....((.((..(((....))((((.((((.........))))))))..)..))))....)))))))))..))))))). ( -32.16) >consensus CGUGCAUAAAAUGCUGAGCACAAAAAAAUUUCCACUGCAUACGUAAUGAGCCUCGAUGAUUCCCAGACCAGAAUAAAAUCCUGGCUGGACGGGGCCAGAAAGUGGCAUUUAAAUGCGCGC (((((((.(((((((..(((...............)))..((....((.(((((((....))((((.((((.........))))))))..)))))))....)))))))))..))))))). (-35.96 = -36.21 + 0.25)

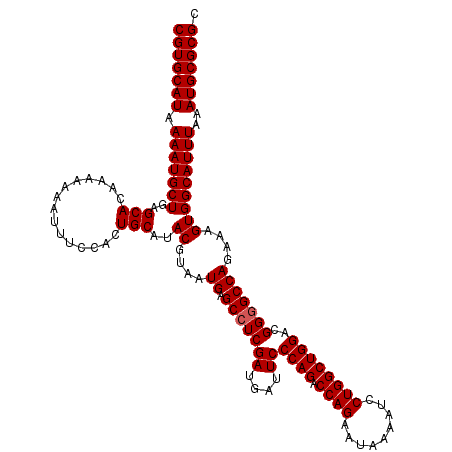
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:40 2006