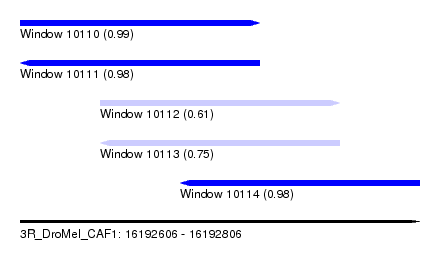
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 16,192,606 – 16,192,806 |
| Length | 200 |
| Max. P | 0.985972 |

| Location | 16,192,606 – 16,192,726 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.26 |
| Mean single sequence MFE | -32.58 |
| Consensus MFE | -29.83 |
| Energy contribution | -30.43 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.985972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16192606 120 + 27905053 UUUCACUGCGGCCUAAUAAAGUAUAAGUAUAGUAUAGGCCAUAUUUGUGGAGGGGAAAACUGGCUGGAGACAGUGGUCCAUAAUAUAUCAUUGCCAUUGUCAAGCUUUGUCUUCCAUUUG .........((((((.((..(((....)))..))))))))......((((((((.((....((((...(((((((((...............))))))))).)))))).))))))))... ( -35.06) >DroSec_CAF1 21905 120 + 1 UUUCACUGCGGCCUAAUAAAGUAUAAGUAUAGUAUAGGCCAUAUUUGUGGAGGGGAACAUCGGCUAGAGACAGUGGUCCAUAAUAUAUCAUUGCCAUUGUCAAGCUUUGUCUUCCAUUUU .........((((((.((..(((....)))..))))))))......((((((((.((....((((...(((((((((...............))))))))).)))))).))))))))... ( -33.96) >DroSim_CAF1 15432 120 + 1 UUUCACUGCGGCCUAAUAAAGUAUAAGUAUAGUAUAGGCCAUAUUUGUGGAGGGGAAAACUGGCUGGAGACAGUGGUCCAUAAUAUAUCAUUGCCAUUGUCAAGCUUUGUCUUCCAUUUG .........((((((.((..(((....)))..))))))))......((((((((.((....((((...(((((((((...............))))))))).)))))).))))))))... ( -35.06) >DroEre_CAF1 21165 101 + 1 ------UGCGGCCUAAUAU-------------UUUCGGCCAUAUUUGAGGAGGGGAAAACUGGCUGCAGACAGUGGUCCAUAAUAUAUCAUUGCCAUUGUCAAGCUUUGUCUUCCAUUUG ------...((((.((...-------------.)).))))........((((((.((....((((...(((((((((...............))))))))).)))))).))))))..... ( -30.76) >DroYak_CAF1 22581 106 + 1 UUUCA-UACGCCCUAAUAU-------------AGUAGGCCAUAUUUGUGGAGGGGAAAACUGGCUGCAGACAGUGGUCCAUAAUAUAUCAUUGCCAUUGUCAAGCUUUGUCUUCCAUUUG .....-...(.((((....-------------..)))).)......((((((((.((....((((...(((((((((...............))))))))).)))))).))))))))... ( -28.06) >consensus UUUCACUGCGGCCUAAUAAAGUAUAAGUAUAGUAUAGGCCAUAUUUGUGGAGGGGAAAACUGGCUGGAGACAGUGGUCCAUAAUAUAUCAUUGCCAUUGUCAAGCUUUGUCUUCCAUUUG .........((((((...................))))))......((((((((.((....((((...(((((((((...............))))))))).)))))).))))))))... (-29.83 = -30.43 + 0.60)



| Location | 16,192,606 – 16,192,726 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.26 |
| Mean single sequence MFE | -27.79 |
| Consensus MFE | -24.01 |
| Energy contribution | -24.81 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982370 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16192606 120 - 27905053 CAAAUGGAAGACAAAGCUUGACAAUGGCAAUGAUAUAUUAUGGACCACUGUCUCCAGCCAGUUUUCCCCUCCACAAAUAUGGCCUAUACUAUACUUAUACUUUAUUAGGCCGCAGUGAAA .....(((((((...(((.((((.((((.((((....)))).).))).))))...)))..)))))))....(((.....(((((((...................)))))))..)))... ( -29.01) >DroSec_CAF1 21905 120 - 1 AAAAUGGAAGACAAAGCUUGACAAUGGCAAUGAUAUAUUAUGGACCACUGUCUCUAGCCGAUGUUCCCCUCCACAAAUAUGGCCUAUACUAUACUUAUACUUUAUUAGGCCGCAGUGAAA .....((..((((..(((.((((.((((.((((....)))).).))).))))...)))...))))..))..(((.....(((((((...................)))))))..)))... ( -26.21) >DroSim_CAF1 15432 120 - 1 CAAAUGGAAGACAAAGCUUGACAAUGGCAAUGAUAUAUUAUGGACCACUGUCUCCAGCCAGUUUUCCCCUCCACAAAUAUGGCCUAUACUAUACUUAUACUUUAUUAGGCCGCAGUGAAA .....(((((((...(((.((((.((((.((((....)))).).))).))))...)))..)))))))....(((.....(((((((...................)))))))..)))... ( -29.01) >DroEre_CAF1 21165 101 - 1 CAAAUGGAAGACAAAGCUUGACAAUGGCAAUGAUAUAUUAUGGACCACUGUCUGCAGCCAGUUUUCCCCUCCUCAAAUAUGGCCGAAA-------------AUAUUAGGCCGCA------ .....(((((((...(((.((((.((((.((((....)))).).))).))))...)))..))))))).............((((....-------------......))))...------ ( -26.40) >DroYak_CAF1 22581 106 - 1 CAAAUGGAAGACAAAGCUUGACAAUGGCAAUGAUAUAUUAUGGACCACUGUCUGCAGCCAGUUUUCCCCUCCACAAAUAUGGCCUACU-------------AUAUUAGGGCGUA-UGAAA .....(((((((...(((.((((.((((.((((....)))).).))).))))...)))..))))))).........(((((.((((..-------------....)))).))))-).... ( -28.30) >consensus CAAAUGGAAGACAAAGCUUGACAAUGGCAAUGAUAUAUUAUGGACCACUGUCUCCAGCCAGUUUUCCCCUCCACAAAUAUGGCCUAUACUAUACUUAUACUUUAUUAGGCCGCAGUGAAA .....(((((((...(((.((((.((((.((((....)))).).))).))))...)))..)))))))............(((((((...................)))))))........ (-24.01 = -24.81 + 0.80)


| Location | 16,192,646 – 16,192,766 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -28.44 |
| Consensus MFE | -27.34 |
| Energy contribution | -27.38 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.614902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16192646 120 + 27905053 AUAUUUGUGGAGGGGAAAACUGGCUGGAGACAGUGGUCCAUAAUAUAUCAUUGCCAUUGUCAAGCUUUGUCUUCCAUUUGCGACUUACAUUUCCCGCAUAUUGUGUUAUUUUUCCGUUUC ........(((((((((.((.((((...(((((((((...............))))))))).))))..)).))))...((((............))))............)))))..... ( -29.16) >DroSec_CAF1 21945 120 + 1 AUAUUUGUGGAGGGGAACAUCGGCUAGAGACAGUGGUCCAUAAUAUAUCAUUGCCAUUGUCAAGCUUUGUCUUCCAUUUUCGACUUACAUUUCCCGCAUAUUGUGUUAUUUUUCCGUUUC ........(((((((((....((((...(((((((((...............))))))))).))))..(((..........))).....))))))((((...))))......)))..... ( -25.36) >DroSim_CAF1 15472 120 + 1 AUAUUUGUGGAGGGGAAAACUGGCUGGAGACAGUGGUCCAUAAUAUAUCAUUGCCAUUGUCAAGCUUUGUCUUCCAUUUGCGACUUACAUUUCCCGCAUAUUGUGUUAUUUUUCCGUUUC ........(((((((((.((.((((...(((((((((...............))))))))).))))..)).))))...((((............))))............)))))..... ( -29.16) >DroEre_CAF1 21186 120 + 1 AUAUUUGAGGAGGGGAAAACUGGCUGCAGACAGUGGUCCAUAAUAUAUCAUUGCCAUUGUCAAGCUUUGUCUUCCAUUUGCGACUUACAUUUCCCGCAUAUUGUGUUAUUUUUCCGUUUC ........(((((((((.((.((((...(((((((((...............))))))))).))))..)).))))...((((............))))............)))))..... ( -29.36) >DroYak_CAF1 22607 120 + 1 AUAUUUGUGGAGGGGAAAACUGGCUGCAGACAGUGGUCCAUAAUAUAUCAUUGCCAUUGUCAAGCUUUGUCUUCCAUUUGCGACUUACAUUUCCCGCAUAUUGUGUUAUUUUUCCGUUUC ........(((((((((.((.((((...(((((((((...............))))))))).))))..)).))))...((((............))))............)))))..... ( -29.16) >consensus AUAUUUGUGGAGGGGAAAACUGGCUGGAGACAGUGGUCCAUAAUAUAUCAUUGCCAUUGUCAAGCUUUGUCUUCCAUUUGCGACUUACAUUUCCCGCAUAUUGUGUUAUUUUUCCGUUUC ........(((((((((.((.((((...(((((((((...............))))))))).))))..)).))))...((((............))))............)))))..... (-27.34 = -27.38 + 0.04)



| Location | 16,192,646 – 16,192,766 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -25.42 |
| Consensus MFE | -23.64 |
| Energy contribution | -24.24 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.745653 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16192646 120 - 27905053 GAAACGGAAAAAUAACACAAUAUGCGGGAAAUGUAAGUCGCAAAUGGAAGACAAAGCUUGACAAUGGCAAUGAUAUAUUAUGGACCACUGUCUCCAGCCAGUUUUCCCCUCCACAAAUAU .....(((..............(((((..........)))))...(((((((...(((.((((.((((.((((....)))).).))).))))...)))..)))))))..)))........ ( -26.30) >DroSec_CAF1 21945 120 - 1 GAAACGGAAAAAUAACACAAUAUGCGGGAAAUGUAAGUCGAAAAUGGAAGACAAAGCUUGACAAUGGCAAUGAUAUAUUAUGGACCACUGUCUCUAGCCGAUGUUCCCCUCCACAAAUAU .....(((...(((......)))..(((((......((((....((.....))..(((.((((.((((.((((....)))).).))).))))...))))))).))))).)))........ ( -22.40) >DroSim_CAF1 15472 120 - 1 GAAACGGAAAAAUAACACAAUAUGCGGGAAAUGUAAGUCGCAAAUGGAAGACAAAGCUUGACAAUGGCAAUGAUAUAUUAUGGACCACUGUCUCCAGCCAGUUUUCCCCUCCACAAAUAU .....(((..............(((((..........)))))...(((((((...(((.((((.((((.((((....)))).).))).))))...)))..)))))))..)))........ ( -26.30) >DroEre_CAF1 21186 120 - 1 GAAACGGAAAAAUAACACAAUAUGCGGGAAAUGUAAGUCGCAAAUGGAAGACAAAGCUUGACAAUGGCAAUGAUAUAUUAUGGACCACUGUCUGCAGCCAGUUUUCCCCUCCUCAAAUAU .....(((..............(((((..........)))))...(((((((...(((.((((.((((.((((....)))).).))).))))...)))..)))))))..)))........ ( -25.80) >DroYak_CAF1 22607 120 - 1 GAAACGGAAAAAUAACACAAUAUGCGGGAAAUGUAAGUCGCAAAUGGAAGACAAAGCUUGACAAUGGCAAUGAUAUAUUAUGGACCACUGUCUGCAGCCAGUUUUCCCCUCCACAAAUAU .....(((..............(((((..........)))))...(((((((...(((.((((.((((.((((....)))).).))).))))...)))..)))))))..)))........ ( -26.30) >consensus GAAACGGAAAAAUAACACAAUAUGCGGGAAAUGUAAGUCGCAAAUGGAAGACAAAGCUUGACAAUGGCAAUGAUAUAUUAUGGACCACUGUCUCCAGCCAGUUUUCCCCUCCACAAAUAU .....(((..............(((((..........)))))...(((((((...(((.((((.((((.((((....)))).).))).))))...)))..)))))))..)))........ (-23.64 = -24.24 + 0.60)



| Location | 16,192,686 – 16,192,806 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.33 |
| Mean single sequence MFE | -21.06 |
| Consensus MFE | -21.00 |
| Energy contribution | -21.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.79 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.983841 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 16192686 120 - 27905053 AAAUGUUCGUUGCCACAACAAAUCAAAAAACGUAAGGCCAGAAACGGAAAAAUAACACAAUAUGCGGGAAAUGUAAGUCGCAAAUGGAAGACAAAGCUUGACAAUGGCAAUGAUAUAUUA .((((((((((((((...............(....).((......))...............((((((...(((...((.......))..)))...)))).)).))))))))).))))). ( -21.00) >DroSec_CAF1 21985 120 - 1 AAAUGUUCGUUGCCACAACAAAUCAAAAAACGUAAGGCCAGAAACGGAAAAAUAACACAAUAUGCGGGAAAUGUAAGUCGAAAAUGGAAGACAAAGCUUGACAAUGGCAAUGAUAUAUUA .((((((((((((((..(((..((......(....).((......))....................))..)))..(((((...((.....))....)))))..))))))))).))))). ( -21.30) >DroSim_CAF1 15512 120 - 1 AAAUGUUCGUUGCCACAACAAAUCAAAAAACGUAAGGCCAGAAACGGAAAAAUAACACAAUAUGCGGGAAAUGUAAGUCGCAAAUGGAAGACAAAGCUUGACAAUGGCAAUGAUAUAUUA .((((((((((((((...............(....).((......))...............((((((...(((...((.......))..)))...)))).)).))))))))).))))). ( -21.00) >DroEre_CAF1 21226 120 - 1 AAAUGUUCGUUGCCACAACAAAUCAAAAAACGUAAGGCCAGAAACGGAAAAAUAACACAAUAUGCGGGAAAUGUAAGUCGCAAAUGGAAGACAAAGCUUGACAAUGGCAAUGAUAUAUUA .((((((((((((((...............(....).((......))...............((((((...(((...((.......))..)))...)))).)).))))))))).))))). ( -21.00) >DroYak_CAF1 22647 120 - 1 AAAUGUUCGUUGCCACAACAAAUCAAAAAACAUAAGGCCAGAAACGGAAAAAUAACACAAUAUGCGGGAAAUGUAAGUCGCAAAUGGAAGACAAAGCUUGACAAUGGCAAUGAUAUAUUA .((((((((((((((......................((......))...............((((((...(((...((.......))..)))...)))).)).))))))))).))))). ( -21.00) >consensus AAAUGUUCGUUGCCACAACAAAUCAAAAAACGUAAGGCCAGAAACGGAAAAAUAACACAAUAUGCGGGAAAUGUAAGUCGCAAAUGGAAGACAAAGCUUGACAAUGGCAAUGAUAUAUUA .((((((((((((((......................((......))...............((((((...(((...((.......))..)))...)))).)).))))))))).))))). (-21.00 = -21.00 + 0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:24 2006